DeBias: generalized framework for analysis of coupled cell biological variables
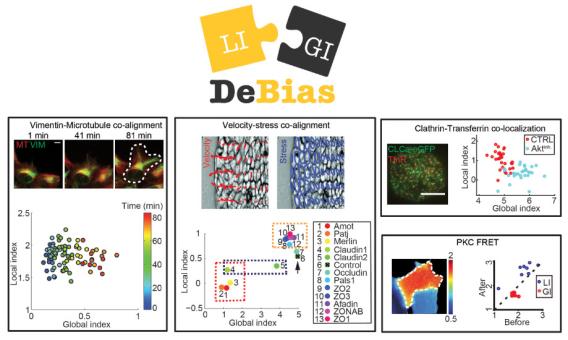
Assaf Zaritsky and colleagues introduced DeBias, software for the analysis of complex relations between coupled cellular variables, published this month in eLife. DeBias decouples the global bias of external effectors from direct interaction, proposing a second measurement for co-localization and co-alignment analyses. Global bias is often a neglected in interpretation of coupled variables, but encapsulates fundamental mechanistic insight into cellular behavior, as showcased in the paper in four different areas of cell biology. The DeBias software package is freely accessible online via the UTSW BioHPC web-server.
eLife is a peer-reviewed open access journal that publishes outstanding fundamental and theoretical research in the life sciences and biomedicine. Led by editor-in-chief Randy Schekman, 2013 Nobel Laureate in Physiology, the journal is one part of a unique, non-profit collaboration between research practitioners and the journal’s funders – Howard Hughes Medical Institute, Max-Planck Society and Wellcome Trust.
Mohan awarded NIH M.D./Ph.D. Fellowship
Congratulations to Ashwathi (Abbee) Mohan on her recently awarded M.D./Ph.D. Fellowship from the National Institutes of Health. These fellowships address the critical need for clinician scientists with both medical training and research experience, to tackle the most complex problems of human health. Abbee’s grant, entitled The mechanical and biochemical mechanism of the oncogenic Rac1P29S mutation in melanoma survival signaling, will enable her to study a hyperactivating melanoma mutation in the Rac1 protein, which coordinates intracellular and extracellular signaling. Melanoma is a highly aggressive malignancy responsible for over 75% of all skin cancer deaths. This research may lead to promising strategies for melanoma drug development that target mechanisms of cancer cell interaction with the surrounding environment.