Econometrics meets molecular cell biology
How can we study molecular processes that are regulated by multiple components and functionally connected to one another through feedbacks? A perturbation may or may not generate a phenotype, but the phenotype, if detectable, is uninterpretable in terms of the function the perturbed component assumes in the unperturbed system – finding that function is the ultimate goal of a cell biological experiment. The phenotype merely indicates how the molecular process adapts to the component dysfunction that is introduced by the perturbation. Our lab has been invested for now more than 15 years in developing a work-around to this fundamental flaw in molecular cell biology.
Jungsik Noh, Tadamoto Isogai and co-authors, along with generations of graduate students and postdocs who have contributed tools and intellectual frameworks before, have finally broken through this wall. They borrowed a statistical tool from another field of science that is concerned with the inference of causal relations in redundant and nonlinear circuitry: econometrics. Contrary to molecular cell biologists, econometricians cannot perturb the regulatory processes under scrutiny. They cannot stop stock trading in Frankfurt, London, and Singapore to infer how the fluctuations in these markets with functionally redundant inputs to Wall Street affect the trading in New York. Econometrists have devised several approaches to infer causal chains from observational, i.e. perturbation-free, data. Among the best known approaches is the concept of Granger-causal inference which prescribes a statistical test to determine the causal influence of a time-resolved variable A on another time-resolved variable B. The concept is straightforward to understand yet is challenging to implement for molecular processes that are controlled in space and time.
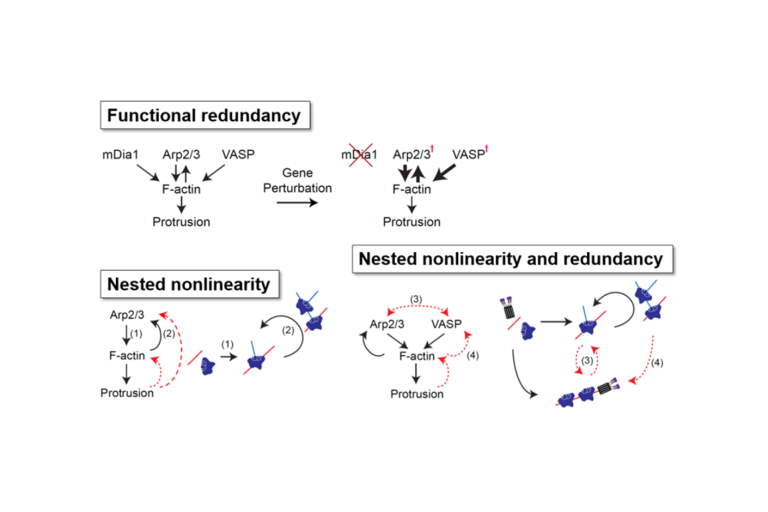
Jungsik and Tada closely collaborated on optimizing the numerical procedures and the experimental conditions to apply perturbation-free causal inference of the circuitry of molecular regulators that govern the dynamics of actin assembly in the lamellipodium, a region at the periphery of cells where actin polymers generate forces that control cell shape. Published in Cell Systems, they applied Granger-causal inference to several hotly-debated questions in the actin cytoskeleton field such as 1) where and how much is actin assembly regulated by the nucleators mDia1, Arp2/3, and VASP? Perturbing one of these will immediately trigger a compensation response that obscures an answer to the original question. 2) It is well-documented that mechanical tension in the protruding cell membrane feeds back on to the assembly of actin filaments (F-actin). However, does this feedback invoke activation of the regulator Arp2/3 or other regulators, and how does feedback from filament assembly to regulator recruitment contribute in parallel? None of these questions can be addressed by perturbation of system components.
Finally, Jungsik and Tada also applied their causal inference pipeline to scrutinize the nonlinear and redundant interactions between Arp2/3, VASP, and F-actin. Quite unexpected, their analysis shows that Arp2/3- and VASP-regulated F-actin structures operate independently, yet synergistically to promote cell protrusion. We believe this study is at the beginning of a paradigm shift in the deep mechanistic analysis of multifactorial molecular systems. Software and further information beyond the paper can be downloaded from our Github website.
 Causal Inference when perturbations fail
Causal Inference when perturbations fail