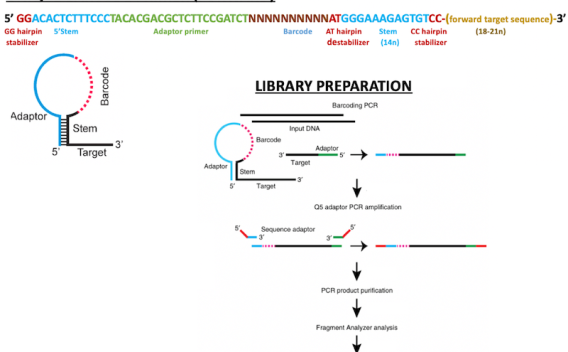
Next-generation sequencing (NGS) is widely used in cancer research and has been a boon to basic and translational research because of its ability to identify low prevalence mutations in heterogenous samples. However, detecting sequence variants below 1% frequency remains a challenge with standard NGS protocols because of the background noise introduced by polymerases during library construction.
In the current research project, we have used an improved library construction approach to mitigate the above concern by using reduced primer concentration, elongated PCR extension times, and hairpin-protected barcode primers to enable simple, multiplexed, PCR-based barcoding of DNA for sensitive mutation detection using sequencing (SiMSen-Seq).
Detecting cell-free DNA from blood plasma may be a potential cancer biomarker. Hence, we have used ctDNA from blood plasma of patients with head and neck squamous cell carcinoma (HNSCC) to monitor the expression of 39 target primers that would be able to detect >95% of invasive HNSCC cases. This analysis can be used as a prognostic tool especially for patients who are negative to HPV16 and also to monitor recurrence after radiotherapy, thus allowing for better prognostication and/or modification of treatment strategies.