
September, 2023
When Cells Catch the Measles
Mazloom-Farsibaf et al. introduced a computational framework to assess the molecular organizations at the cell membrane in a shape-agnostic manner.
View Article
March, 2023
Gone without Blebs!
Conversion of raw light-sheet microscope data to 3D surfaces using u-Shape3D. Poorly adhered MV3 cell embedded in soft bovine collagen. Septins visualized with mouse SEPT6–GFP probe.
View Article
September, 2022
Different Places, Different Shapes
Segal et al. determines environmental cell shape adaptation of human Ewing sarcoma xenografts in zebrafish embryos.
View Article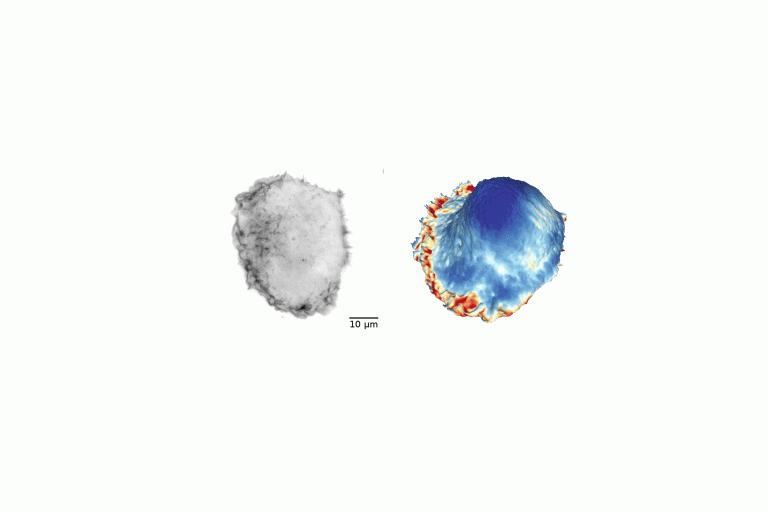
August, 2022
Mixed signals driving oncogenesis: clues from cell morphology
Dr. Gabriel Muhire Gihana awarded the highly competitive HHMI Hanna Gray Fellowship to study how cell morphology regulates RAS oncogenic signaling.
View Article
June, 2022
Econometrics meets molecular cell biology
Jungsik Noh and Tadamoto Isogai implement Granger-causal inference to determine the functional hierarchy of multiple and nonlinearly connected regulators of lamellipodial actin regulator dynamics.
View Article
March, 2022
Dr. Benjamin Nanes awarded a Physician Scientist Career Development Award
The Dermatology Foundation, which funds innovative research in skin biology with the potential to lead to new treatments and cures, has awarded Benjamin Nanes a Physician Scientist Career Development Award. Dr. Nanes is a practicing dermatologist and joined the Danuser laboratory in 2019 to conduct research into the mechanics behind skin remodeling during wound repair or in diseases.
View Article
November, 2021
Danuser Lab takes the lead in launching an innovative metastasis imaging Center
The National Cancer Institute (NCI) at NIH funds innovative research directions in cancer biology. The Cellular Cancer Biology Imaging Research (CCBIR) is one such program that brings together diverse scientists from academic and clinical backgrounds to establish interdisciplinary Centers.
View Article
July, 2021
Mechanism for interpreting complex cell image features driving deep learning classifiers
Deep learning has emerged as the technique of choice for identifying subtle patterns in biomedical data sets, which are meaningful for the classification of the data content.
View Article
April, 2021
SH3BP4 -- the link between adhesion and Akt signaling
In a paper published in the April issue of Developmental Cell, Christoph Burckhardt describes his discovery of the mechanism by which SH3BP4 mediated in an Akt-dependent fashion endocytosis of α5-integrin and neuroplin-1.
View Article
January, 2021
Welf et al. discover the necessity of membrane-actin bond release for cell protrusion initiation
In a collaborative effort, Welf et al. discovered that separation of the cell membrane from an existing actin network is required to initiate the molecular events that lead to cell shape change.
View Article
December, 2020
Gabriel Muhire Gihana Receives the Damon Runyon Fellowship
The Damon Runyon Cancer Research Foundation awards competitive fellowships to selected researchers and physician scientists.
View Article
May, 2020
Jin Suk Park 2020 Nominata Award Recipient
Danuser Lab alum and recent graduate from the UTSW Cancer Biology Graduate Program is the 2020 recipient of the Nominata Award, the highest honor for academic and research accomplishment bestowed by the graduate school to an advanced graduate student.
View Article
April, 2020
Disassembly Asymmetry Score Classification (DASC)
Xinxin Wang and colleagues publish their novel unsupervised classification for endocytosis in eLife.
View Article
February, 2020
Jin Suk Park et. al. discovered a mechanism by which epithelial cells adjust glucose metabolism to extracellular matrix stiffness
Recently published work by Park, Burkhardt, Isogai, Minna, DeBerardinis and Danuser was featured as a Nature News and Views article on February 12, 2020 and as a Research Highlight in Nature Reviews Molecular Cell Biology on February 14, 2020.
View ArticleDecember, 2019
Assaf Zaritsky et. al. deep learning program featured in The Scientist
Recently published work by Zaritsky, Jamieson, Cillay, Nevarez, Welf and Danuser was featured as the "Image of the Day" in an article on December 9, 2019 in The Scientist. The article covers a presentation, Live cell histology for classification of melanoma cell population based on single cell actions (Abstract MS98, page 342), which was given on December 10, 2019 at the combined American Society of Cell Biology and European Molecular Biology Organization 2019 Meeting.
View Article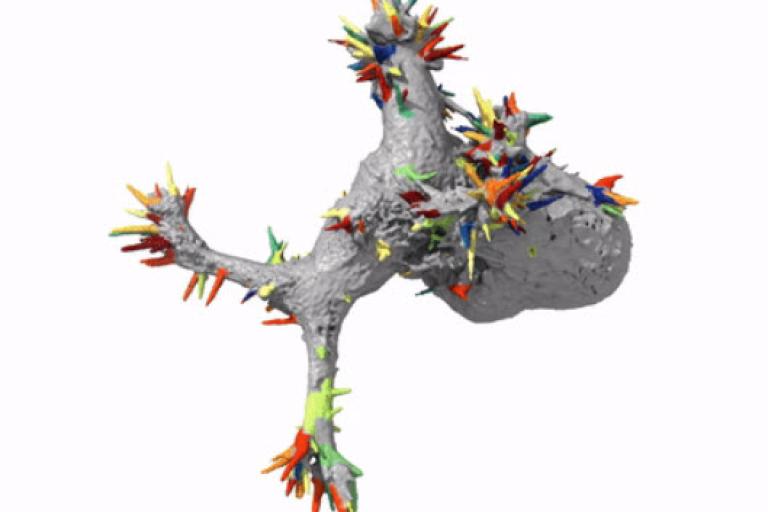
October, 2019
Automatic detection of morphological motifs
Driscoll et al. published an article in the October issue of Nature Methods describing u-shape3D, a suite of computational tools to investigate how 3D cellular morphology governs intracellular signaling. In this article, we introduce a generic morphological motif detector that uses machine learning to find morphological structures, such as lamillipodia, blebs, and filopodia, given user provided examples of these structures. Combining this detector with tools to measure signaling near the cell surface, boundary motion, and other metrics, we measure how Kras and PIP2, two central signaling molecules, associate with blebs, a type of morphological motif. A Behind the Paper blog post details our motivation for building an automated analysis framework for 3D images.
View Article
August, 2019
Discrete events of microtubule depolymerization correlate with continuous activity of GEF-H1, as shown by Poisson point process theory
Azoitei, Noh et al. published a paper in the August issue of the Journal of Cell Biology determining how local changes in microtubule (MT) cytoskeleton regulate the activity of guanine nucleotide exchange factor H1 (GEF-H1).
View Article
May, 2019
The hyperactivating Rac1P29S mutation drives drug tolerance and metastasis in melanoma
A lab team orchestrated by Ashwathi (Abbee) Mohan discovered that the molecular infrastructure that allows a cell to control its shape and migrate can also be engaged in cancer cells to increase signals that promote growth.
View ArticleMarch, 2019
Assaf Zaritsky et. al. deep learning program featured in The Jerusalem Post
Recently published work by Zaritsky, Jamieson, Cillay, Nevarez, Welf and Danuser was the feature of an article on February 25, 2019 in The Jerusalem Post. The article covers a presentation, Live cell histology for classification of melanoma cell population based on single cell actions (Abstract P1894, page 699), which was given on December 10, 2018 at the combined American Society of Cell Biology and European Molecular Biology Organization 2018 Meeting.
View ArticleFebruary, 2019
Release of Growth Cone Analyzer: A new method to quantify morphological variation among complex cellular architectures.
Growth cones are complex, motile structures at the tip of an outgrowing neurite. They often exhibit a high density of filopodia (thin actin bundles), which complicates the unbiased quantification of their morphologies by software. Contemporary image processing methods require extensive tuning of segmentation parameters and are often not sufficiently adaptable to capture morphology changes associated with switches in regulatory signals.
In a study recently published in the Journal of Cell Biology, Bagonis et al. devise a method to overcome such limitations. The robustness of this new tool enabled the study of morphological variation and its relation to motility in both an unperturbed system and in the context of modified Rho GTPase signaling.
The project was initiated jointly with the lab of Dr. Olivier Pertz at the University of Bern.
The Journal of Cell Biology, published by The Rockefeller University Press, is the leading peer-reviewed journal for cell biology.
January, 2019
EMBO Long-Term Research Fellowship awarded to Dagan Segal
Dagan Segal has become an EMBO Long-Term Research Fellow starting January 2019. The EMBO fellowship is a prestigious European 2-year fellowship, with a strong emphasis on international exchange.
Dagan’s work focuses on development of zebrafish and in vitro models to study interactions of cells with their complex surroundings. Joining UT Southwestern in February 2018, Dagan is a joint postdoctoral fellow with the Amatruda and Danuser labs and is combining imaging, genomics, molecular biology, and analytical tools to gain mechanistic understanding of the responses of cells to their environment, particularly in pediatric cancer.
In a study recently published in the Journal of Cell Biology, Bagonis et al. devise a method to overcome such limitations. The robustness of this new tool enabled the study of morphological variation and its relation to motility in both an unperturbed system and in the context of modified Rho GTPase signaling.
The project was initiated jointly with the lab of Dr. Olivier Pertz at the University of Bern.
The Journal of Cell Biology, published by The Rockefeller University Press, is the leading peer-reviewed journal for cell biology.
December, 2018
Assaf Zaritsky et. al. deep learning program featured in The Scientist
A recent presentation of work by Zaritsky, Jamieson, Cillay, Nevarez, Welf and Danuser was highlighted on December 12, 2018 as the Image of the Day in The Scientist magazine. The presentation, Live cell histology for classification of melanoma cell population based on single cell actions (Abstract P1894, page 699), was given on December 10, 2018 at the combined American Society of Cell Biology and European Molecular Biology Organization 2018 Meeting.
Dagan’s work focuses on development of zebrafish and in vitro models to study interactions of cells with their complex surroundings. Joining UT Southwestern in February 2018, Dagan is a joint postdoctoral fellow with the Amatruda and Danuser labs and is combining imaging, genomics, molecular biology, and analytical tools to gain mechanistic understanding of the responses of cells to their environment, particularly in pediatric cancer.
In a study recently published in the Journal of Cell Biology, Bagonis et al. devise a method to overcome such limitations. The robustness of this new tool enabled the study of morphological variation and its relation to motility in both an unperturbed system and in the context of modified Rho GTPase signaling.
The project was initiated jointly with the lab of Dr. Olivier Pertz at the University of Bern.
The Journal of Cell Biology, published by The Rockefeller University Press, is the leading peer-reviewed journal for cell biology.
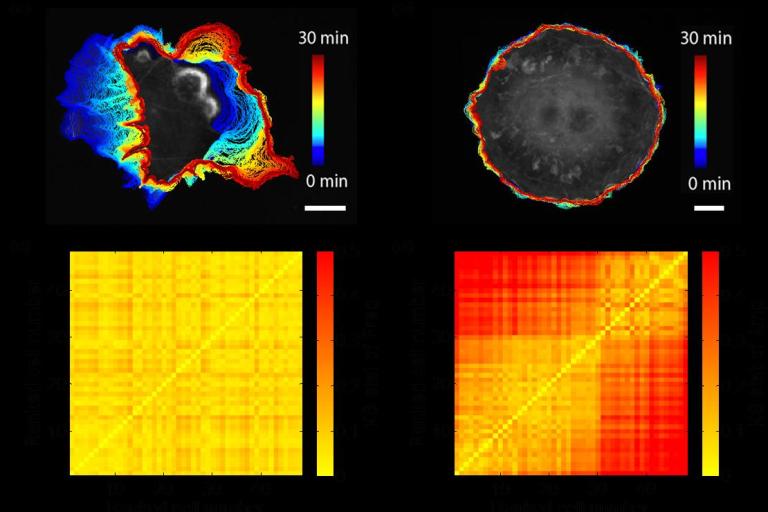
October, 2018
Xiao Ma et. al. PLOS Computational Biology article featured in Science Trends
A paper by postdoctoral fellow Xiao Ma recently published in PLOS Computational Biology, Profiling cellular morphodynamics by spatiotemporal spectrum decomposition (Xiao Ma, Onur Dagliyan, Klaus M. Hahn, and Gaudenz Danuser,) was selected for highlighting in the form of a perspective article by Science Trends, which reports and shares the up-to-date outstanding scientific research progress for a large and global audience.Dagan’s work focuses on development of zebrafish and in vitro models to study interactions of cells with their complex surroundings. Joining UT Southwestern in February 2018, Dagan is a joint postdoctoral fellow with the Amatruda and Danuser labs and is combining imaging, genomics, molecular biology, and analytical tools to gain mechanistic understanding of the responses of cells to their environment, particularly in pediatric cancer.
In a study recently published in the Journal of Cell Biology, Bagonis et al. devise a method to overcome such limitations. The robustness of this new tool enabled the study of morphological variation and its relation to motility in both an unperturbed system and in the context of modified Rho GTPase signaling.
The project was initiated jointly with the lab of Dr. Olivier Pertz at the University of Bern.
The Journal of Cell Biology, published by The Rockefeller University Press, is the leading peer-reviewed journal for cell biology.
May, 2018
Jane Coffin Childs Memorial Fund Fellowship Award
Andrew Weems was recently selected as a Fellow of the Jane Coffin Childs Memorial Fund for Medical Research. One of the most prestigious fellowships available to postdoctoral biomedical researchers, the fund provides three years of support for the brightest young cancer researchers at a critical stage of career growth. Several past Fellows have gone on to become Nobel laureates in physiology, medicine and chemistry.
View ArticleApril, 2018
Human Frontier Science Program Fellowship Award
Jaewon Huh joined the Danuser lab as a postdoctoral fellow in January 2017. This month he was awarded a three-year postdoctoral fellowship from the Human Frontier Science Program (HFSP). The award will provide over $160,000 to support Huh's research that applies financial mathematics to molecular cell biology, creating new frameworks to decipher signal transduction.
View ArticleMarch, 2018
NIH Pathway to Independence Award
Postdoctoral researcher Meghan Driscoll has been awarded a Pathway to Independence Award (K99/R00) from the National Institutes of Health. The grant is designed to facilitate the transition of outstanding researchers from mentored, postdoctoral research positions to independent, tenure-track or equivalent faculty positions. The award will provide over $900,000 over a five-year period to support Driscoll's research program that focuses on how cells migrate through and interact with complex 3D environments.
View ArticleOctober, 2017
Outstanding Educator Award
Gaudenz Danuser, Ph.D. is one of two professors who received 2016-17 Outstanding Educator Awards from the UT Southwestern Academy of Teachers (SWAT) this week in recognition of their teaching excellence in the Graduate School of Biomedical Sciences. The announcement was made, with congratulations, by Dr. Andrew Zinn, Dean of the Graduate School. Dr. Danuser is Professor and Chair of Bioinformatics and member of the faculty of the Cancer Biology and Cell and Molecular Biology graduate programs. A leading researcher in the field of computational and quantitative cell biology, he co-chairs the Computational and Systems Biology track, leads a MatLab programming bootcamp he created for incoming graduate students, has innovated a suite of bioinformatics nanocourses for students and postdocs, and teaches a medical school elective, Mathematical Methods in Medicine.
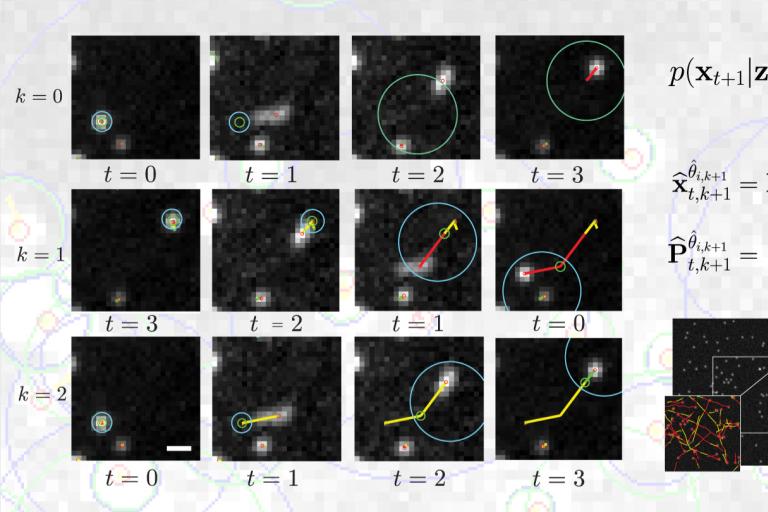
October, 2017
A new model for tracking heterogeneous particle movement in dense environments
A major challenge in the tracking of multiple particle is the capture of heterogeneous movements in crowded scenes. Current models for motion prediction, adapted from aerospatial, are challenged by rapid and unpredictable changes in the nature of the dynamics of objects in motion in a dense intracellular environment. In a study recently published in IEEE Transactions on Image Processing, Roudot et al. propose a piecewise-stationary motion model tailored for intracellular dynamics and a new estimator that exploits multiple tracking rounds within an iterative multiple model smoother (PMMS). This method identifies meaningful dynamical switches in trajectories but also confers an increased robustness toward speed variation and lower acquisition speeds. These results indicate that PMMS is a potent solution to the problem of heterogeneous motion tracking in crowded particle fields which has been a notorious obstacle to high-fidelity analysis of object dynamics, especially in bioimaging applications.
The IEEE Transactions on Image Processing is a leading journal for imaging processing and computer vision. It covers novel theory, algorithms, and architectures for the formation, capture, processing, communication, analysis, and display of images, video, and multidimensional signals in a wide variety of applications.
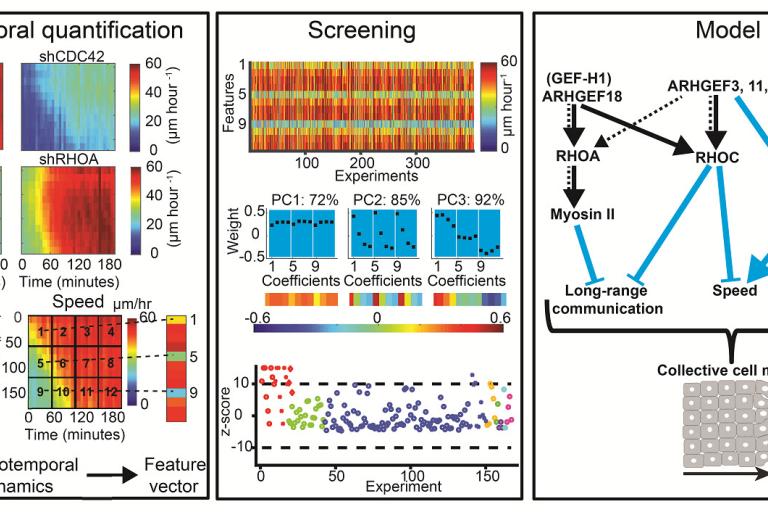
May, 2017
RHOA GEFs regulate long-range intercellular communication during collective cell migration
Effective collective cell migration depends on delicate tuning of cell motility forces and cell–cell communication. Coordination between these processes is regulated, among several pathways, by a family of proteins called the Rho family GTPases. By design of quantitative measures that encode the dynamics of collectively migrating cells in space and time, Assaf Zaritsky, Yun-Yu Tseng, and colleagues identified a surprising role of the Rho GTPases RhoA, RhoC, and their upstream activators in mediating long-range transmission of guidance cues. Their results were published this month in the Journal of Cell Biology.
The project was initiated jointly with Alan Hall, the founder of the Rho GTPase signaling field who tragically died in 2015.
The Journal of Cell Biology, published by The Rockefeller University Press, is the leading peer-reviewed journal for cell biology.
The IEEE Transactions on Image Processing is a leading journal for imaging processing and computer vision. It covers novel theory, algorithms, and architectures for the formation, capture, processing, communication, analysis, and display of images, video, and multidimensional signals in a wide variety of applications.

March, 2017
DeBias: generalized framework for analysis of coupled cell biological variables
Assaf Zaritsky and colleagues introduced DeBias, software for the analysis of complex relations between coupled cellular variables, published this month in eLife.
View Article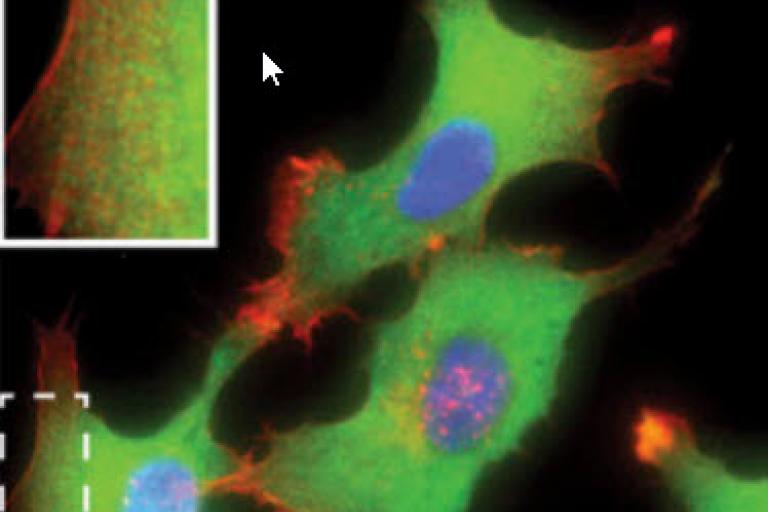
January, 2017
Schafer article journal's most downloaded last year
Congratulations to Claudia Schafer whose article in Convergent Science Physical Oncology (CSPO) was the journal's most downloaded paper for 2015-2016!
View ArticleDecember, 2016
Ning Zhang receives GSO travel award
Congratulations to Ning Zhang, who was selected by the UTSW Graduate Student Organization as one of five Fall Travel Award recipients. Zhang joined the Danuser lab in 2014 as a graduate student in the Cancer Biology program with emphasis in the Computational and Systems Biology track. His research interest is to develop imaging analysis tools for high-throughput 3D spots quantification, which is combined with the fluorescent in situ hybridization (FISH) to analyze genes that can be regulated by telomeres with different lengths. Congratulations Ning!
October, 2016
Danuser Website is Online
Danuser lab website goes online, 3.5 years after the first members opened lab at UTSW and others moved from Harvard Medical School. Over the past few years we have assembled a great team of 20+ researchers with a broad range of scientific backgrounds.