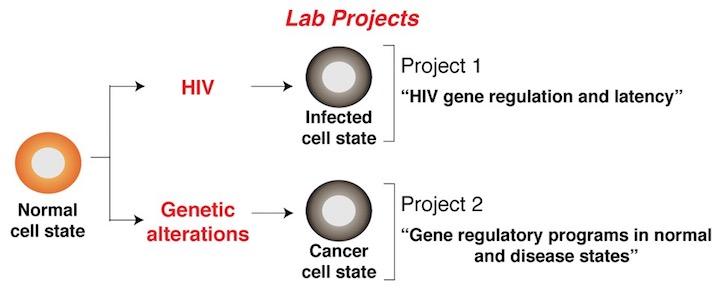
Research in the D'Orso Lab focuses on understanding molecular mechanisms of transcriptional regulation and defining how these normal programs become deregulated during viral infections and disease states. Another aspect is on the host-pathogen interaction between viruses and human cells at the molecular level, with particular emphasis on signaling and gene expression regulation. By understanding how viruses utilize host resources for their propagation, we hope to understand basic physiological processes as well as pathways that can be targeted for therapies.
Transcriptional programs control key biological processes such as differentiation, development, and cellular responses to environmental cues. Thus, their dysregulation promotes malignancy. In this process, normal cells can acquire one or many genetic alterations that dysregulate the gene expression program for cell survival thereby inducing a “cancer cell state”. As such, cancer cells are addicted to unique transcriptional programs that generate vulnerabilities that can be targeted therapeutically.
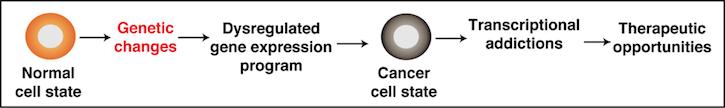
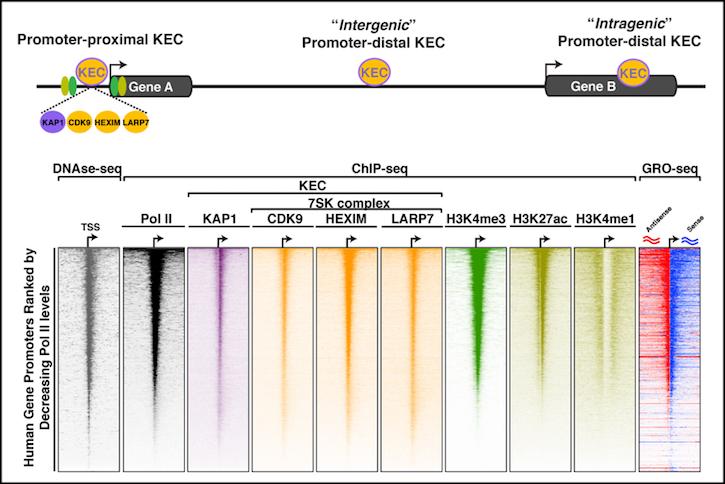
Our lab has recently identified a novel transcription elongation complex referred to as KEC, which controls the expression of thousands of genes (such as growth-driving genes) in cancer cells. Our lab is interested in defining how the KEC operates to regulate transcription to maintain the cancer cell state. At the molecular level, we have recently found that the KEC assembles at both promoters and enhancers in the human genome and that genetic deletion of KEC components dampens transcription of growth-driving genes thereby blocking cell growth. Our lab is currently defining the mechanisms by which the KEC assembles on chromatin through recognition of histone post-translational modifications and sequence-specific DNA elements. Defining the molecular basis of this process will lead us not only to a better understanding of gene regulation but to alternative strategies to target these transcriptional addictions in cancer.
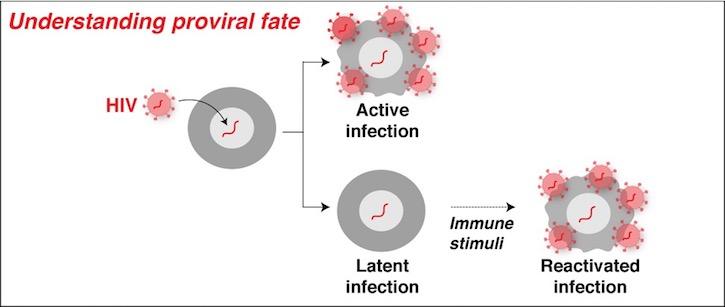
HIV integrates into chromatin-accessible areas of the host genome. The large diversity of the integration landscape suggests that there might be position effects controlling the fate of infection or “proviral fate” (decision-making between active or latent infection). Using both experimental and computational approaches our lab is interested in elucidating hidden regulatory programs (genetic/epigenetic features) that control proviral fate. A better understanding of these regulatory programs will help us devise strategies for personalized medicine approaches to cure patients of HIV infection.
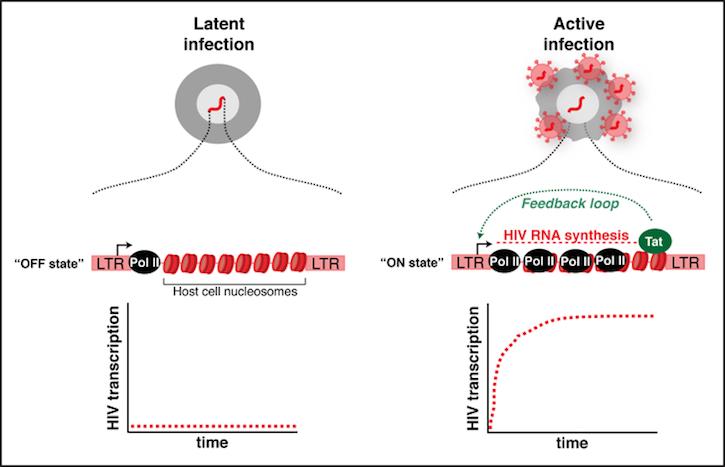
At the molecular level, the dogma in the field poses that during latent infection, the HIV LTR promoter is in an “OFF state” due to RNA polymerase II (Pol II) pausing at the promoter as well as the presence of a restrictive host cell nucleosome barrier at the HIV genome. Conversely, during an active infection, the HIV LTR promoter is in an “ON state” due to the action of cellular activators that promote Pol II pause release thereby leading to the synthesis of the viral-encoded activator Tat, which induces a robust positive feedback loop to sustain HIV transcription over time. However, how the HIV integration site in the host genome dictates proviral fate and what host cell factors control this process remain poorly understood. Unleashing the molecular rules controlling proviral fate will lead us not only to a better understanding of the basic biology of HIV but also will inform us about novel factors and signaling pathways that could be exploited therapeutically.