Research
We study how neural circuits integrate sensorimotor information, memory, and internal state to guide behavior. We use Drosophila as a primary model and employ a multidisciplinary approach that encompasses molecular genetics, neural recording, behavioral experiments and computational modeling, to uncover neural mechanisms that provide animals with behavioral flexibility.
Principal Investigator

Daisuke Hattori, Ph.D.
Assistant Professor | Effie Marie Cain Scholar in Medical Research
Department of Physiology
Department of Neuroscience
Peter O'Donnell Jr. Brain Institute
Daisuke received his bachelor's degree from the University of Tokyo in Japan. His thesis project in Masanori Taira’s lab focused on the early development of the vertebrate brain. For his graduate work in Larry Zipursky's lab at the University of California, Los Angeles, Daisuke studied molecular mechanisms that mediate neural circuit assembly. His work contributed to demonstrating that the diversity encoded by the Drosophila Dscam1 gene is essential to mediate neurite self-avoidance by generating unique neuronal identity labels. As a postdoc in Richard Axel's lab at Columbia University, Daisuke studied the anatomy and function of a neural circuit that supports learning and memory. He contributed to studies that identified a complete circuit diagram of the Drosophila learning center, the mushroom body. In addition, his work revealed a simple dopamine-dependent neural mechanism in Drosophila that mediates the detection of novelty and transition to familiarity. In 2018, Daisuke joined the faculty in the Department of Physiology with a secondary appointment in the Department of Neuroscience.
Current Members
Avirut Mehta
Research Intern
Avirut joined the lab as a summer student and has stayed as a research intern. As the first member of the lab, Avirut has contributed to many projects including establishing different computational pipelines. Avirut earned a BS from UT Dallas with major in International Political Economy and minor in Biology.

Chris Snell
Research Technician
Chris came to the lab through SURF fellowship, then continued as an intern over the next year. He then earned his BS in Neuroscience from UT Dallas and joined the lab as a fulltime technician. In addition to scientific discoveries, Chris enjoys discovering new music and classic horror movies, and understanding current events and global politics.

Gulhumay Gardashova
Grad Student
Gulhumay was born and raised in Turkmenistan and moved to the US for her undergraduate study, graduating from the University of Kansas with BS in Molecular Biology. Gulhumay is currently a PhD student in the Neuroscience Program at UT Southwestern. Outside of lab, Gulhumay spends most of her time pretending to be various action heroes, listening to different iterations of “Baby Shark”, or doing whatever her toddlers command to do that day.

Kristin Connors
Research Assistant
Originally from New Mexico, Kristin earned her BS from Southern Methodist University with a major in Biology and minors in Mechanical Engineering and Chemistry. Kristin joined the lab as a research assistant and, as one of the original members of the lab, has been involved in many different projects including development of new assays.

Rahi Dakwala
Research Technician
Rahi graduated from UT Austin with BS in Neuroscience and joined the lab as a research technician. Outside the lab, Rahi enjoys talking to friends, watching Bollywood movies, and playing basketball.

Ryan McGrath
Research Assistant
Ryan graduated from the University of Illinois Urbana Champaign with a Master of Engineering in Bioengineering, where he also earned his BS in Bioengineering with minor in Molecular and Cellular Biology. In his free time, Ryan likes to work on his personal 3D printer as part of the maker community.
Xinke Wen
Grad Student
Xinke grew up in a small town in Shandong, China, and developed her interest in neuroscience during high school biology class. Xinke went to Wuhan University to pursue her interest and earned her BS in Biological Science. Xinke is currently a PhD student in the Neuroscience Program at UT Southwestern. In her spare time, Xinke reads poems and plays with her cats. She currently focuses on teaching her cat to tell left from right.

Alumni
Alexa De La Torre Schutz
Analia Marzoratti (Green Fellow)
Divya Gollapalli (Green Fellow)
Gabriella Williams (STARS Fellow)
Giann Ruiz (STARS Fellow)
Jack Mostyn (Green Fellow)
Jackie Dominguez (STARS Fellow)
Moise Bonheur
Publications
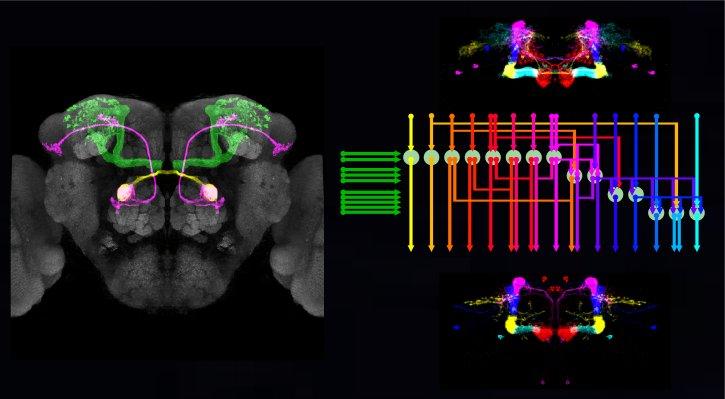
A rapid and bidirectional reporter of neural activity reveals neural correlates of social behaviors in Drosophila.
Bonheur M*, Swartz KJ*, Metcalf MG*, Wen X*, Zhukovskaya A, Mehta A, Connors KE, Barasch JG, Jamieson AR, Martin KC, Axel R, Hattori D. Nature Neuroscience (2023) 26 (7), 1295-1307
A neural theory for counting memories.
Dasgupta S, Hattori D*, Navlakha S*. Nature Communications (2022) 13 (1), 5961
Representations of Novelty and Familiarity in a Mushroom Body Compartment.
Hattori D, Aso Y, Swartz KJ, Rubin GM, Abbott LF, Axel R. Cell (2017) 169 (5), 956-969
The neuronal architecture of the mushroom body provides a logic for associative learning.
Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TT, Dionne H, Abbott LF, Axel R, Tanimoto H, Rubin GM. eLife (2014) 3:e04577
Robust discrimination between self and non-self neurites requires thousands of Dscam1 isoforms.
Hattori D, Chen Y, Matthews BJ, Salwinski L, Sabatti C, Grueber WB, Zipursky SL. Nature (2009) 461 (7264), 644-648
Dscam-mediated cell recognition regulates neural circuit formation.
Hattori D*, Millard SS*, Wojtowicz WM*, Zipursky SL. Annual Review of Cell and Developmental Biology (2008) 24, 597-620, Review
Dscam diversity is essential for neuronal wiring and self-recognition.
Hattori D*, Demir E*, Kim HW, Viragh E, Zipursky SL, Dickson BJ. Nature (2007) 449 (7159), 223-227
Dendrite self-avoidance is controlled by Dscam.
Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB. Cell (2007) 129 (3), 593-604
Got diversity? Wiring the fly brain with Dscam.
Zipursky SL, Wojtowicz WM, Hattori D. Trends in Biochemical Sciences (2006) 31 (10), 581-588, Review
DNA methylation controls the timing of astrogliogenesis through the regulation of JAK-STAT signaling.
Fan G, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, Hattori D, Ge W, Shen Y, Wu H, ten Hoeve J, Shuai K, Sun YE. Development (2005) 132 (15), 3345-3356
Identification of target genes for the Xenopus Hes-related protein XHR1, a prepattern factor specifying the midbrain-hindbrain boundary.
Takada H*, Hattori D*, Kitayama A, Ueno N, Taira M. Developmental Biology (2005) 283 (1), 253-267
Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies.
Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, Chess A, Zipursky SL. Neuron (2004) 43 (5), 673-686
DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation.
Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. Science (2003) 302 (5646), 890-893
Lab Fun
Contact Us
Department of Physiology
UT Southwestern
5323 Harry Hines Blvd
MC9040, ND12.300
Dallas, TX 75390
(214) 645-6060 (office)
(214) 645-7030 (labs)
Email Daisuke