Caspases are thought to be universal effectors of apoptotic cell death. These enzymes are synthesized as dormant 'prozymogens' and, during apoptosis, they are activated during complex cascades of proteolytic activation. These events ultimately mediate cell death by cleaving selected substrates, thereby reorganizing cellular physiology and promoting self-destruction. Apoptosis can be prevented when caspase function is blocked, either by mutation or by viral or human-made inhibitors.
CED-3, a gene discovered in the nematode, represents a founding member of the pro-apoptotic caspase family. CED-4 bears homology to vertebrate APAF-1 and Drosophila Dark. These 'adaptor' proteins activate apical caspase function via a multimeric complex referred to as the 'apoptosome.' In flies and mammals, an emerging picture for apoptosis control is consistent with a 'gas and brake' model, whereby concurrent input from APAF-1/Dark adaptors, together with removal of IAP inhibition drives caspase activation to levels that exceed a threshold necessary for apoptosis.
The balance of opposing regulatory forces determines the status of apical caspase activity, but the relative contribution from positive regulators (APAF-1/Ced-4/Dark) and negative regulators (the IAP family) can vary among different cell types and species. For example, in the worm, mouse, and fly, mutations in positive regulators cause phenotypes defective in global cell death, but this pattern is not consistently true with respect to the negative regulators (IAPs). Therefore, while underlying components are broadly conserved, the regulatory 'linchpins' in different cells and in different species can vary.
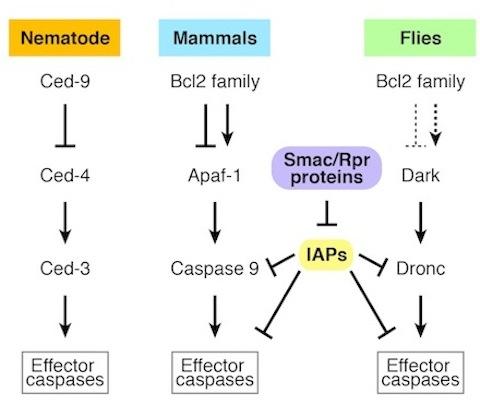
Core apoptotic genes are homologus in nematodes, mammals, and flies.
We exploit a genetic model, Drosophila, to understand the physiology of cell death during normal development and after cell injury. In this animal, deletion of a complex genomic interval, designated the Reaper region, prevents all programmed cell death (PCD).
We discovered four genes in this region that function as pivotal activators of the apoptotic pathway Reaper, Grim, Hid, and Sickle. In germline transformation experiments, these genes partially rescue cell death defects and, in development, these death activators are expressed in patterns that precede the onset of PCD. In cultured cells and in transgenic animals, each gene is sufficient to provoke apoptosis that can be suppressed by caspase inhibitors.
The Reaper genes function through parallel circuits to engage a common set of effectors including caspases. Proteins encoded by the Reaper region function, in part, to liberate active caspases from inhibition by inhibitor-of-apoptosis proteins (IAPs) through activities that are homologous to mammalian IAP antagonists (e.g. Smac/Diablo).
We also discovered another essential regulator of apical caspase activation in flies: Dark. This gene is homologous to human Apaf-1 and to nematode CED-4. Like its mammalian counterpart, the protein is essential for the apoptogenic action of apical caspases and mutations at this gene exhibit profound failures in apoptosis. In related studies, we also established that Dark is a central effector of histolytic cell death and pivotal in some models of pathologic injury, where cell death is provoked by genotoxic stress or polyglutamine toxicity.
Our lab conducted genetic analyses of the two apical caspases in flies: Dronc and Dredd.Dronc encodes the only caspase-recruitment-domain-containing caspase in the Drosophila genome and, as such, exhibits shared structural features with initiator caspases in mammals. Animals lacking Dronc present a range of defects, including extensive hyperplasia of hematopoietic tissues and persisting neuronal cells. Furthermore, in ex vivo preparations, Dronc- cells were completely insensitive to induction of cell killing by diverse stimuli.
Together, these studies place Dronc, and its activator Dark, at an obligate position for converging pathways that specify PCD and stress-induced apoptosis.
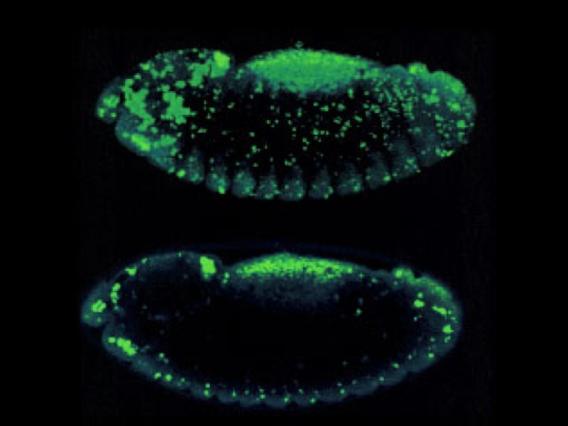
Apoptotic cell death is visualized in a Drosophila embryo using acridine orange. Top: A z-series 'stack.' Bottom: A single confocal 'slice.'
Probing Genes and Networks that Specify Cell Death
The Drosophila apoptosome, like its mammalian counterparts, is a molecular machine that occupies a central position in networks that specify programmed cell death (PCD). We pursue unbiased genetic approaches to gain a comprehensive view of pivotal cell death regulators and to discover novel apoptogenic functions.
Screening for Genes for Programmed Cell Death (PCD) in Vivo
We showed that lesions in canonical apoptotic pathways cause epithelial cells to persist past when they would normally die after eclosion. A uniquely characteristic late-onset phenotype results. Novel PCD genes, if similarly eliminated in the wing, show these same distinctive defects. Therefore, we are conducting a large-scale screening effort and have already recovered several promising mutants. Some mutations encode essential apoptotic determinants in distinct tissue types.
High-Throughput Gene Silencing Platforms
RNAi technology, combined with high-throughput cell-based assay systems, enable methodical testing of annotated genes for functional properties. We are exploiting cell-based RNAi screening platforms to capture novel effectors of apoptotic cell death using stimuli and drugs that simulate programmed signals (e.g. Reaper proteins) or stress from injury.
Stimulus-Induced Apoptosis and Rescue by Gene Silencing
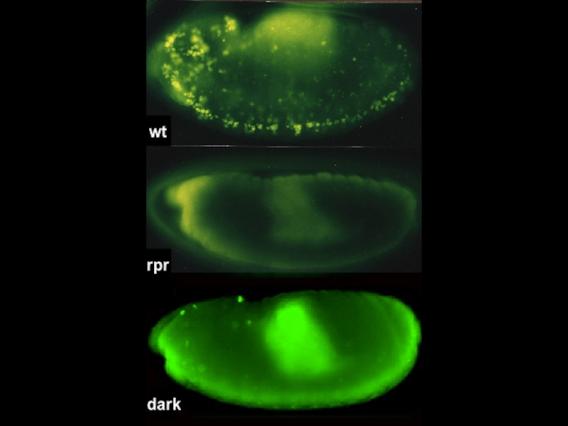
Acridine-stained apoptotic cells are prevalent in the wild-type embryo (top), absent in mutants that lack Rpr proteins, and severely reduced in animals that lack Dark, a component of the fly apoptosome.
Prompted by unique phenotypes associated with cell-death mutations, we developed live reporter systems for real-time imaging of epithelial cell death in vivo. Combining these tools with examination of tissue mosaics for apoptotic activity, we discovered a form of collective cell death, where synchronous apoptosis is coordinated throughout the tissue and causes wholesale loss of an entire epithelium within minutes.
This collective behavior starkly contrasts with typical apoptosis models in developing systems, where a single cell, surrounded by viable neighbors, sporadically initiates apoptosis. Current studies are examining how collective cell death is controlled and coordinated to affect dramatic change as part of tissue remodeling.
Videos
An apoptotic wave progresses through the epithelium of a Drosophila wing in minutes. The top two videos show low magnification. The bottom video shows high magnification.
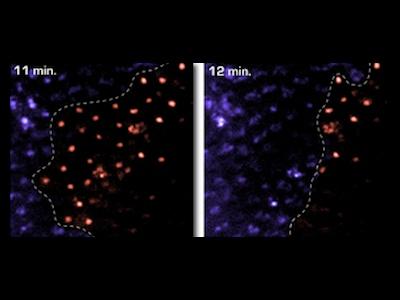
Cell groups die collectively throughout the epithelium, with obvious parallels to glandular involution in mammals.