Tumor Suppression, Cell Death and Mobile Genetic Elements
Abrams Lab applies high throughput genetic approaches to explore two biomedical themes. One organizing theme explores the p53 regulatory network, which is deranged in most human cancers. Despite extensive characterization, precisely how p53 acts to suppress tumors remains poorly understood. We built innovative tools to interrogate p53 function in Drosophila, zebrafish, and mouse models, and, using these, we discovered that p53 tonically acts to suppress transposons. Current projects are directed toward understanding how p53 functions to restrain mobile elements and test the clinical utility of this 'transposopathy' model. A second and related research program examines gene-directed programs that specify programmed and unprogrammed cell death.
Research Areas
In humans, p53 is altered in most human cancers but, despite extensive characterization, precisely how p53 acts to suppress tumors remains poorly understood. By exposing the ancestral properties of p53, we hope to illuminate why this tumor suppressor becomes deranged in human cancers. Toward this objective, we built innovative tools to interrogate p53 function in the Drosophila and zebrafish systems, and, leveraging these, we discovered that p53 tonically acts to suppress transposons.
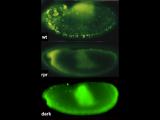
Furthermore, in ‘humanized’ p53 fly strains (that exchange fly p53 for human counterparts) normal human p53 genes contained transposons but mutant alleles arising in cancer patients cannot. Taken together, these discoveries suggest that p53 acts through highly conserved mechanisms to repress transposons and suggest that p53 suppresses oncogenic disease, in part, by restraining mobile elements. Wilms tumor ORF1 stain Wilms tumors mutant for p53 show elevated human LINE1 ORF1p expression.
We believe these insights may help explain why p53 defects are permissive for cancers and might also explain the instability that characterizes most cancer genomes. Consistent with these ideas, we uncovered direct evidence for unrestrained retrotransposons in p53-mutant mouse cancer models and in genotyped patient tumor cancers.
These insights open opportunities to examine the ‘transposopathy’ hypothesis as a general principle in p53 biology.
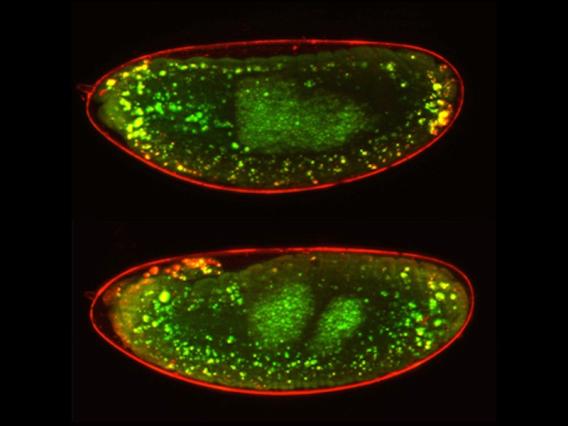
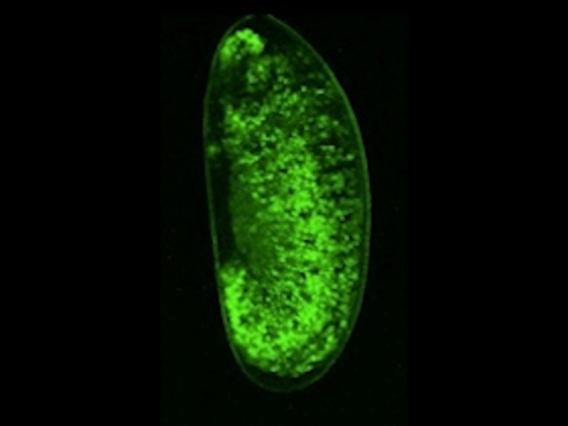
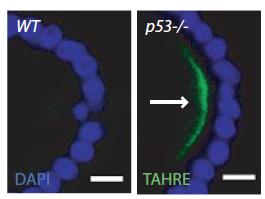 In p53-/- ovaries, TAHRE accumulates in the germ plasm
In p53-/- ovaries, TAHRE accumulates in the germ plasm 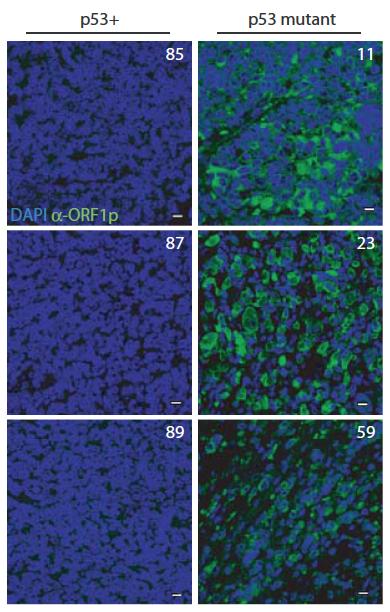 Wilms tumors mutant for p53 show elevated human LINE1 ORF1p expression
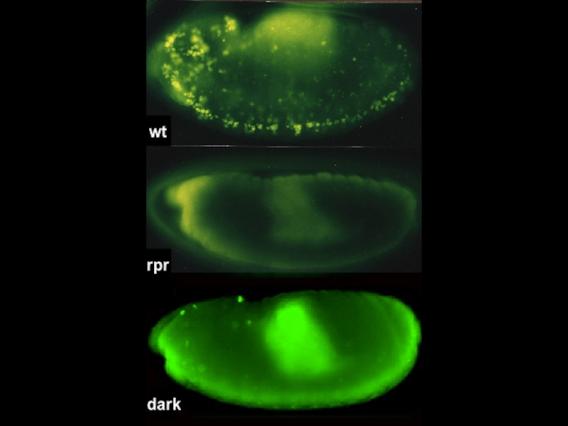
Wilms tumors mutant for p53 show elevated human LINE1 ORF1p expression New technologies in genomic sciences allow us to explore the content of genomes in ways never before possible. While these advances have greatly improved our analytical power, vast portions of the genome that don't encode protein products remain mysterious. The extent of unannotated "non-coding" transcription is not accurately known, but its high activity challenges our concepts of genetic information and of the "junk DNA paradox."
Our research is moving beyond descriptive studies of "genomic dark matter" to determine how "noncanonical" RNAs might encode meaningful content.
p53 genes are widely implicated in age-related diseases. The p53 tumor suppressor gene is mutated in most human cancers. The gene acts to preserve genome stability and constrain oncogenic potential by governing adaptive responses to genotoxic stress.
At the single-cell level, we have a rich body of knowledge about the function of p53. However, little is known about how this gene functions among cell groups to elicit adaptive responses at the tissue level. To illuminate core ancestral functions of this critical tumor suppressor and understand how this gene coordinates injury responses in tissue, we initiated a comprehensive analysis of Dp53, the Drosophila homolog of p53. Like its mammalian counterpart, Dp53 is crucial for genome stability and also for stress-induced apoptotic responses. We further demonstrated that the pro-apoptotic activator, reaper, is an authentic Dp53 target in vivo.
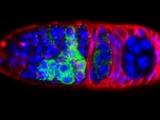
The Dp53/reaper axis contributes important functions during unscheduled apoptosis as an adaptive response to injury. Our research continues to explore conserved properties of adaptive stress responses as they engage the p53 regulatory network. Stained Drosophila embryo p53 is mutated in most human tumors. We recently found that the first catalytic step of genetic recombination during meiosis provokes p53 activity (green), seen here in a fly germarium counterstained for nuclei (blue) and HTS protein (red). From Lu et al. (2010) Science 328: 1278-81. PMID: 20522776.
Using the Drosophila model, we identified a signature profile for p53-dependent stress responses and established a novel method to examine the pro-apoptotic action of p53 in living animals. Together with real-time imaging methods, these tools permit us to examine p53-driven behaviors of living cells in situ.
We are following stimulus-dependent p53 action in live tissues to discover new determinants that specify adaptive responses beyond the single-cell level. From these studies, we found that p53 acts selectively in stem cells.
More recently, we discovered that mutations in p53 genes permit eruptions of mobile elements, known as transposons. We are exploring how p53 contains mobile elements and interrogating pathologic outcomes triggered by unrestrained transposons. At the same time, we are also testing interventions that could mitigate p53-driven ’transposopathies’ by inhibiting the action of these mobile elements.
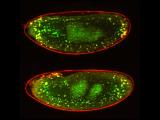
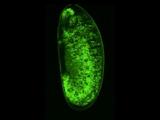
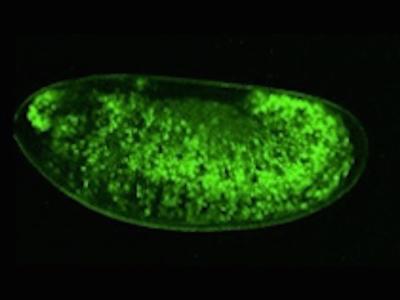 p53 is a well-conserved gene that is mutated or altered in most human cancers. Here, a transgenic reporter produces a green signal when the p53 network is activated by ionizing radiation. This reporter allows us to image Drosophila embryos for native p53 activity in real time.
p53 is a well-conserved gene that is mutated or altered in most human cancers. Here, a transgenic reporter produces a green signal when the p53 network is activated by ionizing radiation. This reporter allows us to image Drosophila embryos for native p53 activity in real time. 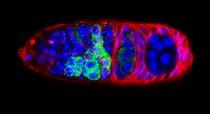 p53 is mutated in most human tumors. We recently found that first catalytic step of genetic recombination during meiosis provokes p53 activity (green), seen here in a fly germarium counterstained for nuclei (blue) and HTS protein (red). From Lu et al. (2010) Science 328: 1278-81. PMID: 20522776.
p53 is mutated in most human tumors. We recently found that first catalytic step of genetic recombination during meiosis provokes p53 activity (green), seen here in a fly germarium counterstained for nuclei (blue) and HTS protein (red). From Lu et al. (2010) Science 328: 1278-81. PMID: 20522776. Our research examines gene-directed programs that specify programmed and unprogrammed cell death. In both vertebrates and invertebrates, dying cells often progress through a series of ultrastructural changes called apoptosis. Stimuli that provoke apoptosis have been extensively investigated in numerous experimental models and it is well established that this form of programmed cell death (PCD) requires genetic functions within the dying cell.
Apoptosis is important not only for development but also as an adaptive response during cellular injury and viral infection. Extensive evidence links aberrant apoptosis to the etiology of cancer, neurodegenerative disorders, auto-immunities, AIDS, and heart disease. The cloning of genes necessary for PCD has spurred our collective understanding of apoptotic cell death and established that core components of the "apoptotic engine" are highly conserved. Despite our advanced body of knowledge, fundamental gaps in our understanding of apoptotic death in biology and medicine remain.
Emerging literature also supports the view that other forms of cell suicide occur and, like apoptosis, these alternate forms of cell death are controlled through genetically encoded programs.
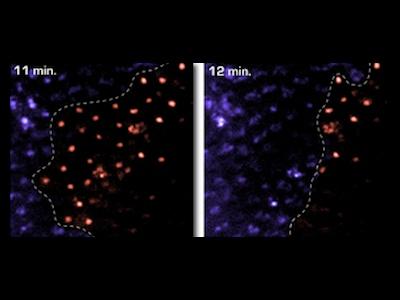
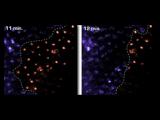
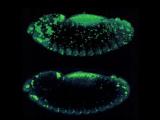
 Apoptotic cell death is visualized in a Drosophila embryo using acridine orange. Top: z-series
Apoptotic cell death is visualized in a Drosophila embryo using acridine orange. Top: z-series As vectors of both parasitic and viral diseases, endemic mosquito populations are a global concern. In Africa, mosquitoes contribute to at least one million deaths each year and, here in the United States, mosquito-borne diseases are also on the rise. Strategies for mitigating the transmission of these diseases require the use of insecticides but conventionally applied agents lack specificity and lose efficacy as resistance inevitably emerges. Consequently, there is an urgent need to develop new agents that are narrowly targeted, environmentally harmless, and safe for humans. Traditional searches for new insecticides involve whole animal screening but, unfortunately, these approaches sample compounds on very limited scales and lack environmental safety tests early in the pipeline. Furthermore, rationale design principles commonly leveraged for drug development programs have not been routinely exploited for pesticide development.
To address these limitations, we recently conducted a pilot study to test the feasibility of an alternative discovery platform. Specifically, we examined whether cell-based screens and counterscreens could be leveraged as a surrogate to identify targeted, vector-specific toxins when tested in whole animals. Towards this goal, we designed a high throughput platform that systematically searches for compounds that are lethal to cultured mosquito cells but show negligible activity against human, rodent, or other insect cell lines.
Our strategy captured a rare set of compounds that specifically killed mosquito cells and, importantly, targeted killing in culture was associated with selective toxicities in whole animal assays.
Furthermore, where tested, strains resistant to a commonly used insecticide (pyrethroid) were also susceptible. Taken together, our results demonstrate that capabilities enabled by this platform dramatically outperform whole animals screens and can expose untapped mosquito-specific vulnerabilities.
We are now capitalizing on this proof-of-concept to deliver the most potent and most selective mosquitocides obtainable from massive chemical and natural product libraries. A successful precedent here could inspire similar efforts to develop customized pesticides that target other disease vectors as well.
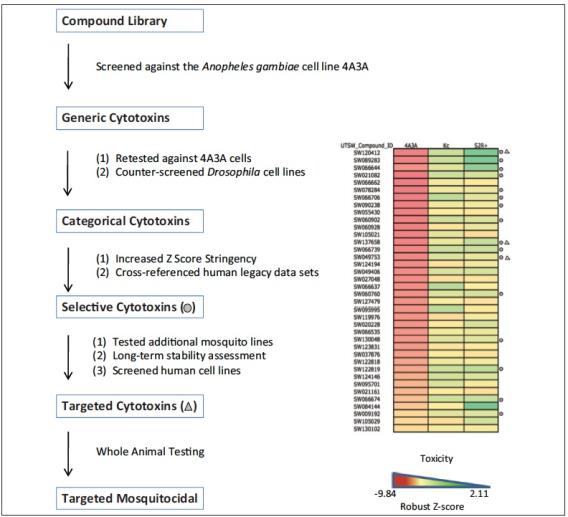 A cell-based screening platform identifies targeted mosquito-specific cytotoxins
A cell-based screening platform identifies targeted mosquito-specific cytotoxins 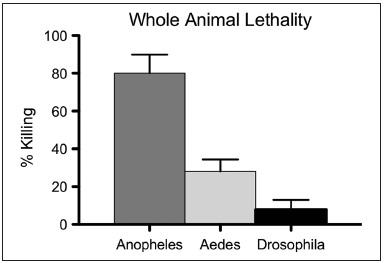 SW120412 showed impressive potency against A. gambiae, whereas Drosophila larvae were unaffected for eclosion rates, even when exposed to 200 μM SW120412
SW120412 showed impressive potency against A. gambiae, whereas Drosophila larvae were unaffected for eclosion rates, even when exposed to 200 μM SW120412 Bioessays: p53 in the Game of Transposons (October 15, 2016)
University Lecture Series (September 14, 2016)
Dr. Abrams was a featured participant in these webinars, sponsored by Science Magazine/AAAS:
The Many Roads to Cell Death (June 14, 2014)
Apoptotic Signaling in Normal and Cancer Cell Biology (September 22, 2009)
Apoptosis Tutorial
This apoptosis tutorial was created by Rebecca Litton, a former student in the medical illustration program.
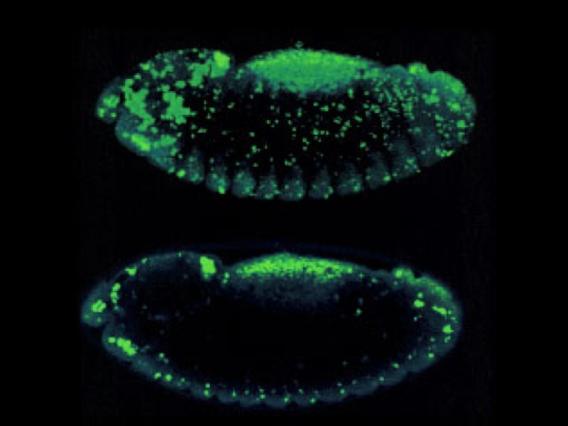
Collective Cell Death
Collective cell death visualized in a Drosophila wing.