Research in the D’Orso lab focuses on understanding how gene regulatory programs operate using a wide array of experimental and computational approaches.
Ongoing projects for incoming trainees (students and postdocs):
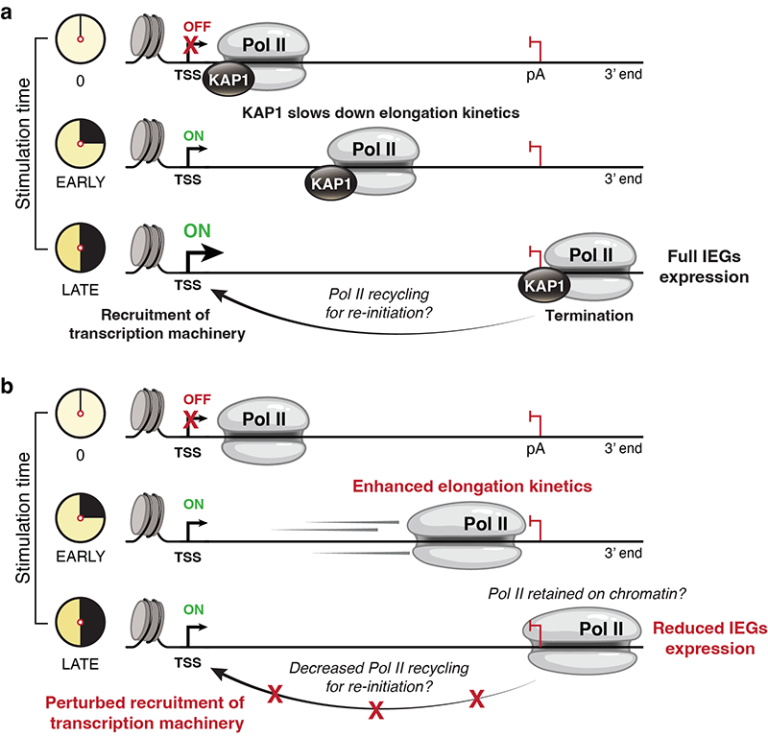
Cell fate decisions are critical for key biological processes including T cell activation and differentiation. These cellular decisions are orchestrated through the precise spatio-temporal activation and deactivation of cell signaling–transcription programs facilitating the expression of Immediate Early Genes (IEGs). We have recently described a counterintuitive mechanism whereby the negative regulation of RNA Polymerase (Pol II) elongation facilitates transcription activation during cell stimulation (see Reference below). Ongoing and future studies will reveal how the interplay between heterochromatin formation and transcription elongation coordinates the proper activation of effector programs during immune cell fate choice and dysregulation in diseased states.
References: KAP1 negatively regulates RNA polymerase II elongation kinetics to activate signal-induced transcription
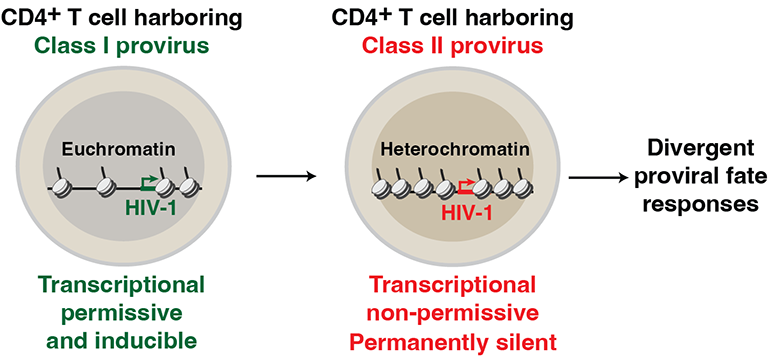
HIV-1 integrates semi-randomly in the human genome, thus generating proviral classes of active and silent genomes. We are interested in defining the mechanisms underlying transcriptional and epigenetic regulation of the HIV-1 proviral classes in immune cells. We want to understand how the euchromatin and heterochromatic genomic environments facilitate the recruitment of certain classes of epigenetic regulators for either HIV-1 proviral transcriptional activation or silencing. Unleashing the molecular rules controlling proviral fate will lead us not only to a better understanding of basic HIV-1 biology, but also inform about novel factors and signaling pathways that could be exploited therapeutically.
References: This is part of an ongoing investigation. Ruess et al., BBI 2022, PMID: 35250265
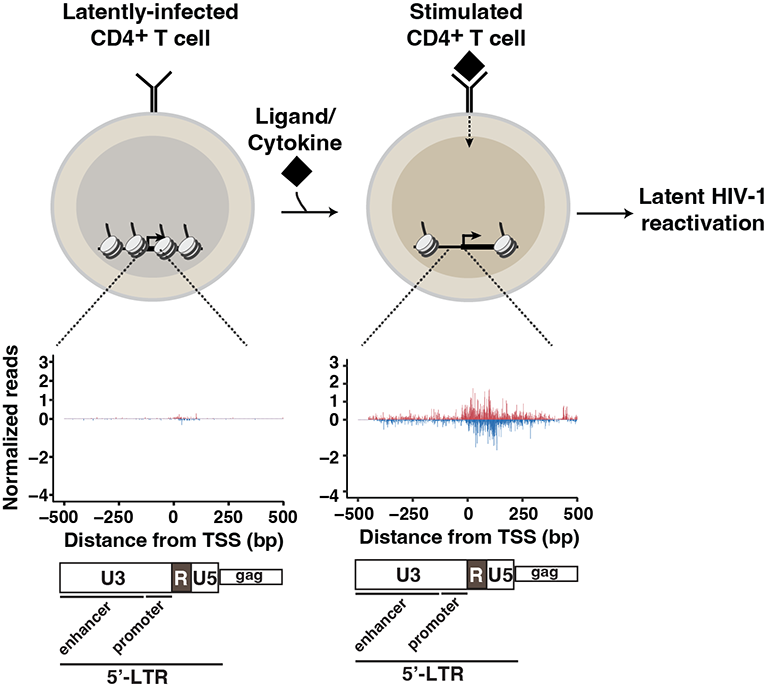
HIV-1 latency and persistence in reservoir cells hamper viral eradication. Defining the mechanisms underlying regulation of latency maintenance and reactivation are paramount for devising alternative cure approaches. Using high-resolution genomics assays, we have gained previously unprecedented knowledge about the HIV-1 transcription process. Ongoing and future studies will reveal how enhancers in the proviral genome, host and viral factors, chromatin and three-dimensional genome architecture collectively coordinate the maintenance of viral latency and reactivation.
References: The HIV-1 Transcriptional Program: From Initiation to Elongation Control and manuscript in preparation.
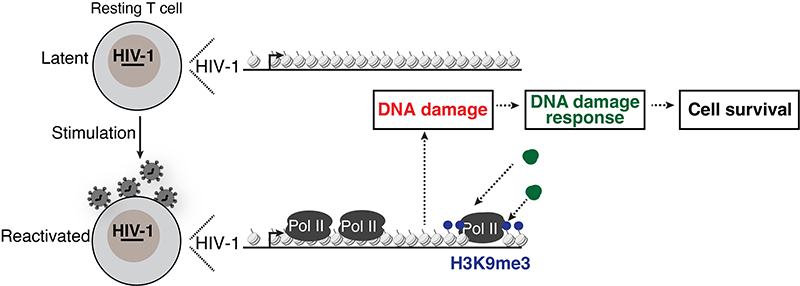
We have recently made the unexpected observation that the process of HIV-1 transcription during latency reactivation induces DNA damage downstream of the provirus and a cellular DNA damage response (DDR). Ongoing and future studies will help define the DDR components involved in the sensing and repair of damaged DNA including nucleic acid structures, cellular factors and chromatin enzymes.
References: This is part of an ongoing investigation.
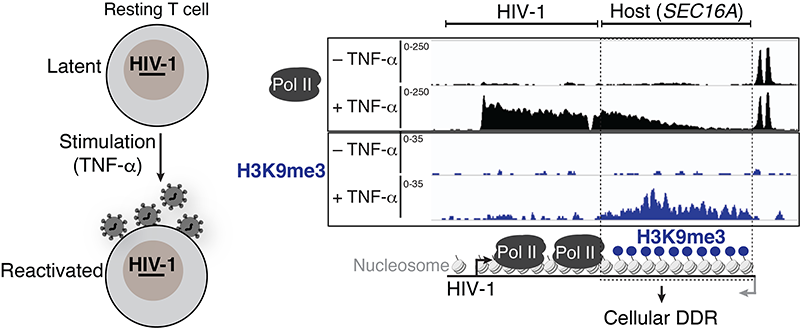
To achieve the goal of eradicating the intact and inducible reservoir, we are leveraging a previously unknown facet in HIV-1 biology for clinical intervention. We have recently made the unexpected observation that the process of HIV-1 transcription during latency reactivation generates localized DNA damage downstream of the provirus (termed here HIV-1 transcription–induced cellular DNA damage). We are targeting major DNA damage and replication stress repair enzymes from the DNA damage response (DDR) during latency reactivation for the selective elimination of cells harboring inducible proviruses over bystander cells and those harboring uninducible proviruses. Our ongoing and future studies will contribute to the daunting task of achieving a functional HIV-1 cure.
References: This is part of an ongoing investigation.