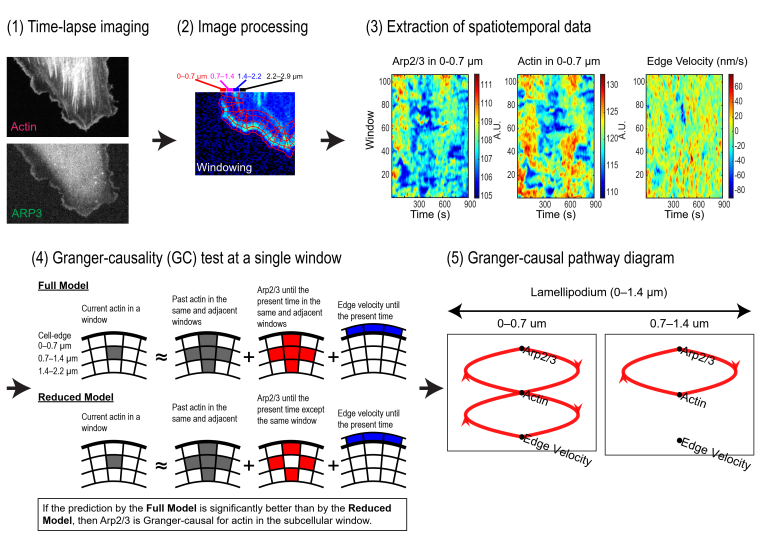
Granger-causal inference of the lamellipodial actin regulator hierarchy
Most cellular processes are regulated by molecular circuitry with functional overlap and nonlinear relation among components. In this configuration the study of individual component functions by experimental intervention is incomplete. We present Granger-Causality analysis applied to observational, high-resolution live cell imaging data as powerful approach to overcome this central challenge in cell biological inquiry. We demonstrate the framework by defining the roles of functionally redundant actin regulators in controlling lamellipodial actin dynamics and cell edge movements.
Our analysis identifies distinct zones of actin regulator activation and of causal effects on filament assembly and delineates actin-dependent and actin-independent regulator roles in controlling edge motion. We propose that edge motion is driven by assembly of two independently operating actin filament systems.
DynamicNeuronTracker (DyNT) segments and tracks jittering single neurons in 3D imaging of deforming tissues
Calcium fluorescence imaging enables us to investigate how individual neurons of live animals encode sensory input or drive specific behaviors. Extracting and interpreting large-scale neuronal activity from imaging data are crucial steps in harnessing this information. A significant challenge arises from uncorrectable tissue deformation, which disrupts the effectiveness of existing neuron segmentation methods. Here, we propose an open-source software, DynamicNeuronTracker (DyNT), which generates dynamic neuron masks for deforming and/or incompletely registered 3D calcium imaging data using patch-matching iterations. We demonstrate that DyNT accurately tracks densely populated neurons, whereas a widely used static segmentation method often produces erroneous masks. DyNT also includes automated statistical analyses for interpreting neuronal responses to multiple sequential stimuli.
We applied DyNT to analyze the responses of pheromone-sensing neurons in mice to controlled stimulation. We found that four bile acids and four sulfated steroids activated 15 subpopulations of sensory neurons with distinct combinatorial response profiles, revealing a strong bias toward detecting sulfated estrogen and pregnanolone.
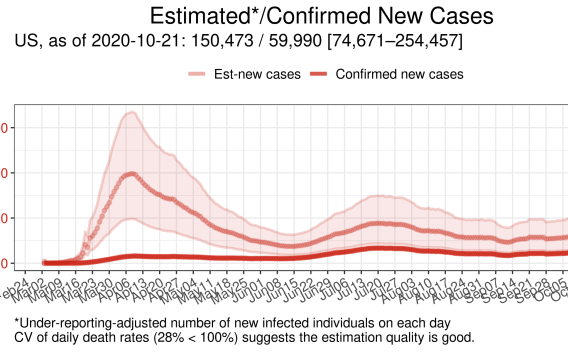
Estimation of daily ascertainment rates of COVID-19 cases unveils actual sizes of currently infected populations
Since the beginning of the coronavirus disease 2019 (COVID-19) pandemic, daily counts of confirmed cases and deaths have been publicly reported in real-time to control the virus spread. However, substantial undocumented infections have obscured the true size of the currently infected population, which is arguably the most critical number for public health policy decisions. We developed a machine learning framework to estimate time courses of actual new COVID-19 cases and current infections in all 50 U.S. states and the 50 most infected countries from reported test results and deaths. Using published epidemiological parameters, our algorithm optimized slowly varying daily ascertainment rates and a time course of currently infected cases each day.
Severe under-ascertainment of COVID-19 cases was found to be universal across U.S. states and countries worldwide. In 25 out of the 50 countries, actual cumulative cases were estimated to be 5–20 times greater than the confirmed cases, as of September 3, 2020. Our estimates of cumulative incidence were in line with the existing seroprevalence rates in 46 U.S. states. In the U.S. states like Louisiana, Georgia, and Florida, more than 4% of the population was estimated to be currently infected, as of September 3, 2020, while in New York this fraction is 0.12%. The estimation of the actual fraction of currently infected people is crucial for any definition of public health policies, which up to this point may have been misguided by the reliance on confirmed cases.