Our Research
The broad research interest of my lab is in dissecting molecular mechanisms of essential membrane-associated cellular events in eukaryotic cell development.
Autophagy is a lysosomal degradation pathway. Numerous human diseases, such as cancer, and infectious and developmental diseases have been linked to aberrant autophagy. Besides its disease relevance, autophagy is a unique system for investigating the basic principles of organellar biogenesis because, unlike many organelles that are rich in membrane and protein components (endoplasmic reticulum, mitochondria, etc), autophagosomes are relatively simple. Autophagosomal biogenesis responds to a variety of internal and external signals, making fine genetic or chemical manipulation possible for understanding molecular mechanisms and defining new targets for disease treatment. Canonically, nascent autophagosomes fuse with lysosomes to deliver their cargos for degradation. However, under certain circumstances, autophagosomes appear to also have degradation-independent functions, for example, protein secretion. We propose that the canonical and non-canonical roles of autophagosomes co-operate to guide cells through critical stages of development, such as meiosis, when the cell is programmed to undergo drastic cellular content degradation and structural remodeling. Our studies will shed light directly on autophagy functions in eukaryotic gametogenesis, the production of sperm (spermatogenesis) and oocytes (oogenesis), and more broadly, on how the autophagic machinery rearranges membranes for crucial aspects of development.
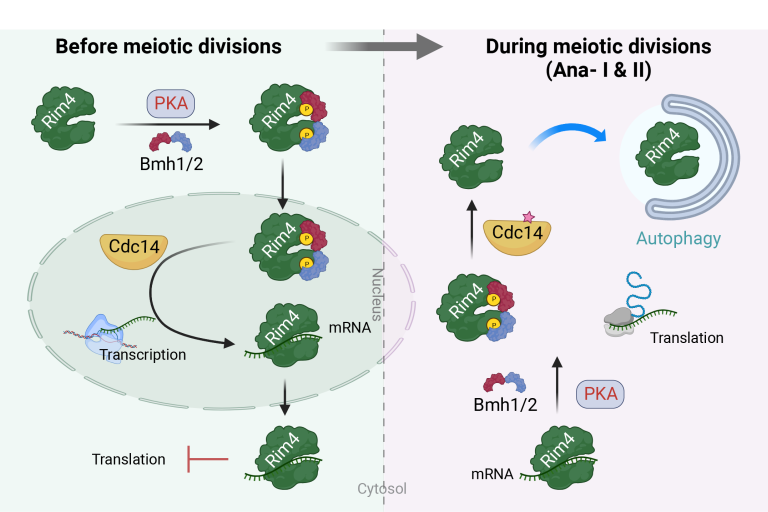 Regulation of Rim4 Distribution, Function, and Stability during Meiosis by PKA, Cdc14, and 14-3-3 Proteins
Regulation of Rim4 Distribution, Function, and Stability during Meiosis by PKA, Cdc14, and 14-3-3 Proteins Proteins arising from small ORFs that are substantially smaller than ~50 residues are considered small proteins (SPs) which historically tend to be ignored. Recent studies, however, suggest that hundreds or even thousands of SPs were synthesized in bacteria and eukaryotes with functions implicated in diverse processes such as spore formation, cell division, movement of molecules across the membrane, enzymatic activity, and signal transduction. SPs are small and prone to interact with their targets, often on the membrane, to modulate their activities, therefore with great potential in clinical application. We are interested in elucidation of a molecular mechanism related to SPs: the targeting machinery, their structure nature, interactions with other molecules as well as their turnover in eukaryotes.
Yeast genetics; Live cell imaging; Electronic microscopy; Proteomic analyses of protein interactomes; Protein biochemistry; Biochemical reconstitution of purified proteins and lipids; in vivo protein labeling
 FM
FM  Lab
Lab Meet the Principal Investigator

Fei Wang, Ph.D.
Fei Wang received his B.S. in Genetics and M.S. in Gene Therapy from Fudan University, China, in 1997 and 2001, respectively. He attended graduate school at UMass-Amherst and earned a Ph.D. in Biochemistry and Molecular Biology with Danny Schnell, Ph.D. He was then trained as a postdoctoral fellow in the research group of Vlad Denic, Ph.D., at Harvard University on defining the molecular mechanism of post-translational translocation of the single pass integral ER membrane proteins: Tail-anchored proteins.
In 2017, Dr. Wang joined the faculty at UT Southwestern in the Center for Autophagy Research, Department of Internal Medicine with a secondary appointment in the Department of Cell Biology.
Meet the Lab Members
Rudian Zhang, Ph.D.
Research Scientist
Research Interests:
Autophagic degradation of protein aggregation/meiotic translational control

Jin Li, Ph.D.
Postdoctoral Researcher
Research Interests: The relationship between SGs (stress granules) and cellular organelles during the progress of meiosis

Former Lab Members
Jayaprakash Monala, Ph.D.
Postdoctoral Researcher

Akshay Chellappa
Research Technician

Featured Publications
Autophagy-Mediated Surveillance of Rim4-mRNA Interaction Safeguards Programmed Meiotic Translation
Rudian Zhang, Wenzhi Feng, Suhong Qian, Fei Wang 2023 Cell Reports in pressCdc14 spatiotemporally dephosphorylates Atg13 to activate autophagy during meiotic divisions.
Feng W, Argüello-Miranda O, Qian S, Wang F, 2022 May J Cell Biol 5 221Autophagy of an Amyloid-like Translational Repressor Regulates Meiotic Exit.
Wang F, Zhang R, Feng W, Tsuchiya D, Ballew O, Li J, Denic V, Lacefield S, 2020 Jan Dev. Cell 2 52 141-151.e5The Get1/2 transmembrane complex is an endoplasmic-reticulum membrane protein insertase.
Wang F, Chan C, Weir NR, Denic V 2014 Aug Nature 7515 512 441-4The mechanism of tail-anchored protein insertion into the ER membrane.
Wang F, Whynot A, Tung M, Denic V 2011 Sep Mol. Cell 5 43 738-50Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Stefer S, Reitz S, Wang F, Wild K, Pang YY, Schwarz D, Bomke J, Hein C, Löhr F, Bernhard F, Denic V, Dötsch V, Sinning I 2011 Aug Science 6043 333 758-62Contact Us
Fei Wang, Ph.D.
Assistant Professor
Department of Cell Biology and Biochemistry
Nancy Cain and Jeffrey A. Marcus Scholar in Medical Research, in Honor of Dr. Bill S. Vowell
Email Dr. Wang
Phone: 214-648-8491
UT Southwestern Medical Center, NL6. 120F
6000 Harry Hines Blvd.
Dallas, TX 75390
Join Our Lab
We are committed to providing a pleasant environment to energetic and motivated researchers interested in science who like to work independently and on a small team. To request a position as a postdoctoral fellow, graduate student, or technician, please contact us.
