RESEARCH OVERVIEW
Overarching Research Interests
In Deja Lab we focus on metabolism and the development of metabolomic and fluxomic approaches to understand metabolic regulation in health and disease.
Our general approach involves:
1) Application of stable isotope tracers to living systems to probe metabolic activity in vivo and ex vivo;
2) Use of analytical chemistry methods such as Nuclear Magnetic Resonance (NMR) spectroscopy and Mass Spectrometry (MS);
3) Utilization of mouse genetics, optogenetics and exercise modalities to gain deeper insight into regulation of metabolism.
4) Development and utilization of variable scale computational models of metabolic pathways;
Metabolic Flux Analysis (MFA): Using stable isotope labeling information in and mathematical models of metabolism we estimate metabolic fluxes.
Biomedical application: By combining in vivo metabolic flux analysis with neurometabolic manipulations we aim to understand how the brain controls liver and whole-body metabolism in obesity and exercise and advance the field of neuro-hepatology.
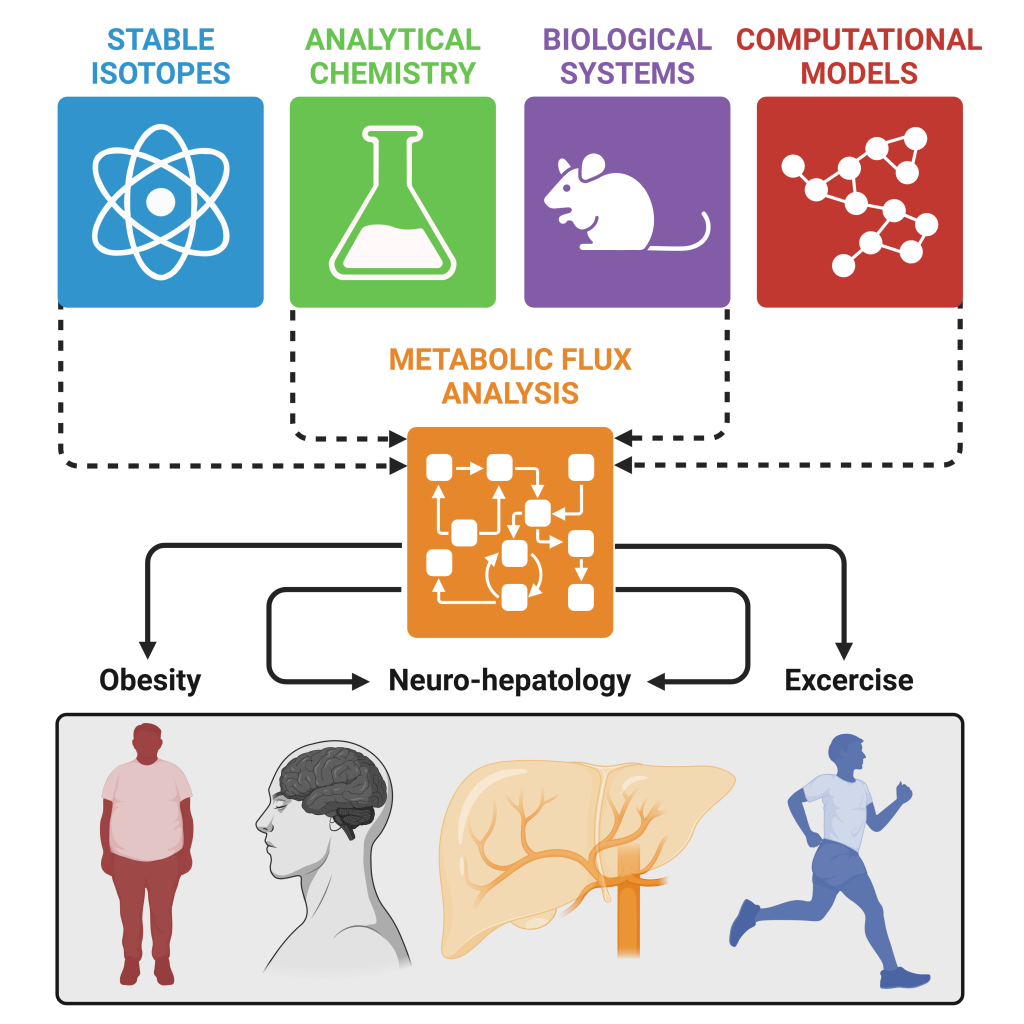 Overview of research areas in Deja Laboratory
Overview of research areas in Deja Laboratory RESEARCH METHODS
1. Tracing metabolic activity using stable isotopes
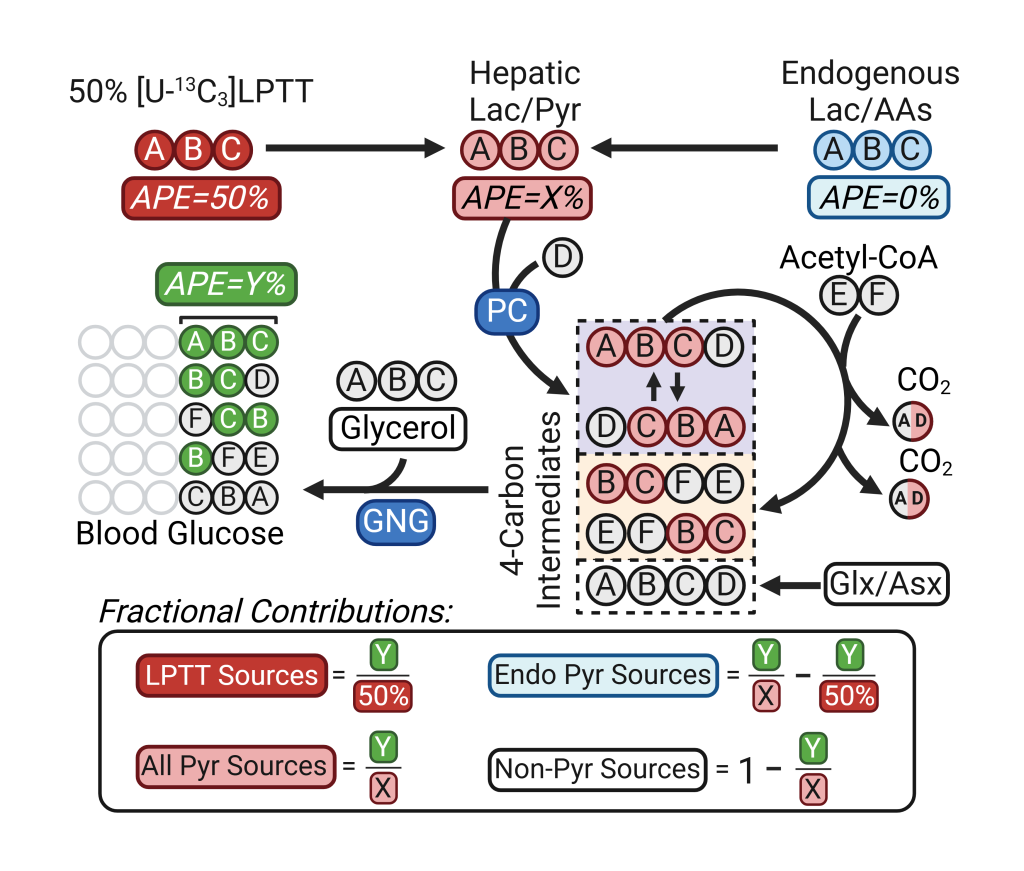 Deja S. et al. Cell Metab . 2024 May 7;36(5):1088-1104.e12. doi: 10.1016/j.cmet.2024.02.004
Deja S. et al. Cell Metab . 2024 May 7;36(5):1088-1104.e12. doi: 10.1016/j.cmet.2024.02.004 The cellular mechanisms of many diseases impact complex metabolic networks that may be targets for therapy if they can be identified. While metabolomics provides static snapshot of metabolism, tracing experiments are required to quantify metabolic flux. Thus, a general approach to characterizing metabolism is to estimate metabolic flux using isotopically labeled substrates (e.g., tracers containing (13C, 2H, 15N, etc.) that incorporate into downstream metabolites (tracees). Information about the pathways that convert the tracer to the tracee is encoded in the number and position of the isotopic nuclei that appear in the tracee. If these labeling patterns can be detected, mathematical models that relate metabolic activity to the formation of these labeling patterns can be used to decode flux through complex networks of metabolism.
We use various stable isotope approaches to monitor metabolism in vitro, ex vivo and in vivo. In the schematic on the left we present a simple workflow for determining 13C carbon contribution to the process. Such analysis provides a useful insight into the substrate partitioning between different pathways using precursor/product proportionality. Mathematical modeling of such data can provide more detailed flux analysis.
2. Analytical methods for monitoring stable isotope labeling in metabolites
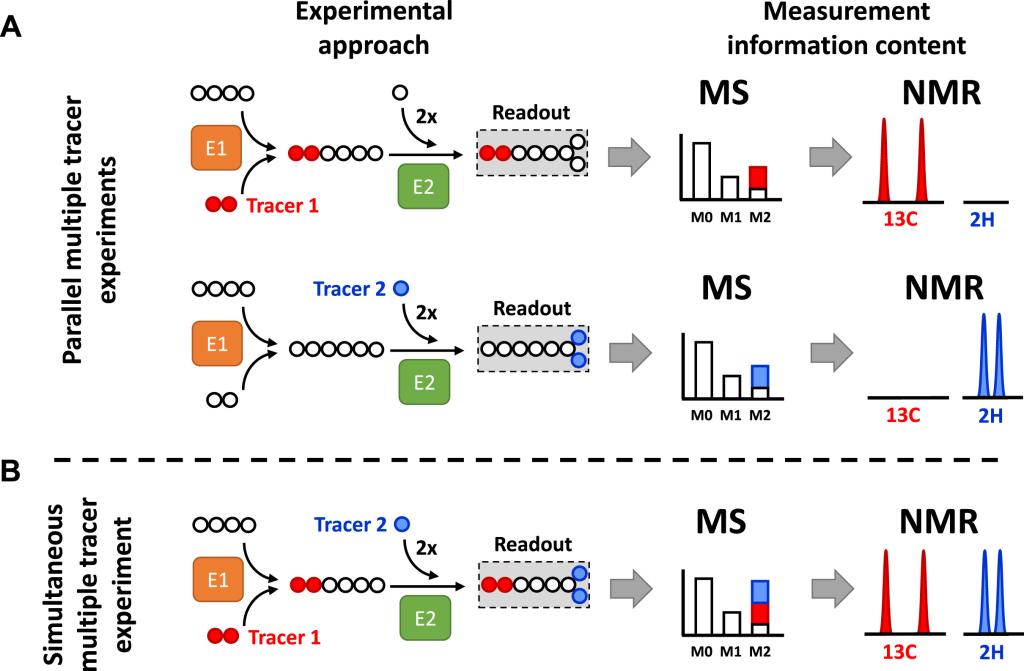 Deja S. et al. Metab Eng . 2020 May:59:1-14. doi: 10.1016/j.ymben.2019.12.005
Deja S. et al. Metab Eng . 2020 May:59:1-14. doi: 10.1016/j.ymben.2019.12.005 We use and develop new methods to measure stable isotope incorporation into metabolites using Nuclear Magnetic Resonance (NMR) spectroscopy and Mass Spectrometry (MS). NMR detects nucleus- and position-specific labeling, while MS detects a shift in mass based on the number of heavy atoms incorporated into analyte creating mass isotopomer distribution (MID). Schematic on the left depicts the application of multiple stable isotope tracers in example metabolic network consisting of two enzymatic reactions (E1 and E2). Tracer 1 provides two 13C atoms, while tracer 2 provides two 2H atoms. (A) Parallel experimental setup: tracers 1 and 2 are administered in two separate biological experiments generating two separate datasets. Therefore, individual MIDs and NMR spectra contain labeling information originating from each single tracer. (B) Simultaneous experimental setup: both tracers are administered in a single biological experiment and generate a single dataset. Labeling from tracer 1 and 2 produces an ambiguous M2 in the MID. In contrast, detection by nucleus-sensitive NMR distinguishes between signals originating from 2H and 13C atoms.
3. Metabolic flux analysis and model development
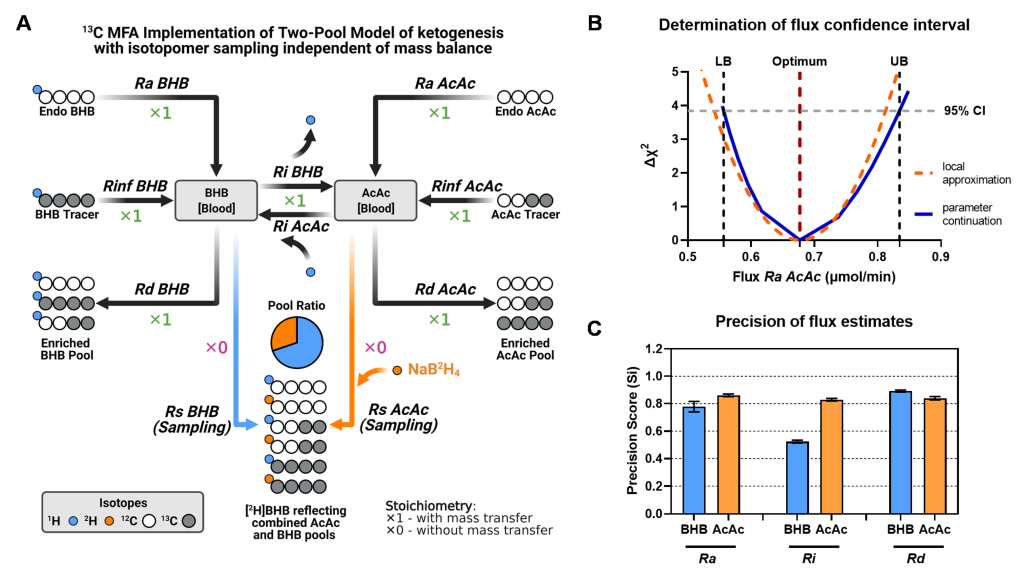 Deja S. et al. Metabolites . 2021 Apr 28;11(5):279. doi: 10.3390/metabo11050279
Deja S. et al. Metabolites . 2021 Apr 28;11(5):279. doi: 10.3390/metabo11050279 We work on development of novel metabolic flux analysis models, and their rigorous validation. For example, the two major analytical methods for flux measurement (NMR and MS) are typically used independent of each other, despite their data being complimentary. We recognize this synergy and created a unified single metabolic model of hepatic metabolism based on mass isotopologue and NMR isotopomer data which included TCA cycle, gluconeogenesis and glycogenolysis, and showed that both NMR and MS, in most cases, highly correlated and responsive to physiological interventions when regressed with an equivalent model This work was accomplished in specialized software tool (INCA 2.0) which we helped to develop. In schematic on the left is an example of MFA model of ketogenesis reliant on a double tracer method. This approach quantifies acetoacetate and β-hydroxybutyrate pool sizes and their rates of appearance, disposal, and exchange while regression analysis provides confidence intervals and detects potential errors in experimental data.
4. In vivo stable isotope infusions
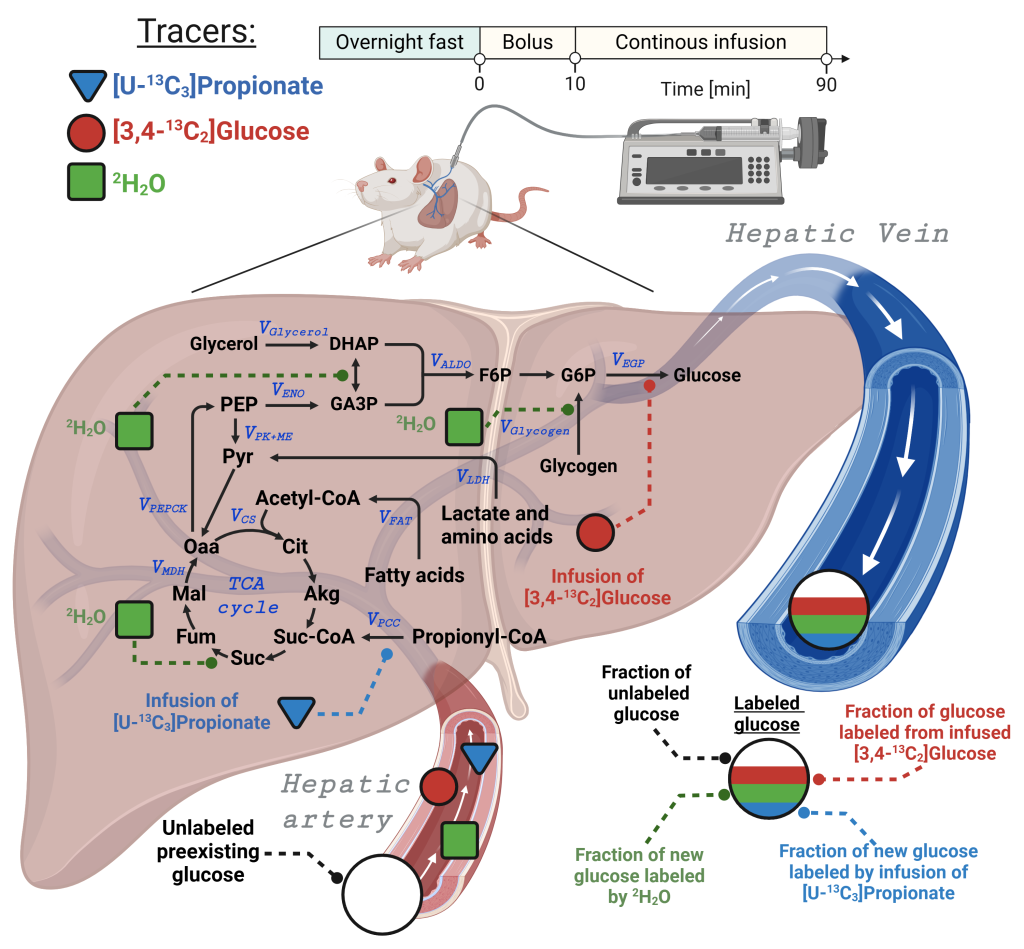 Deja S. et al. Diabetes 2022;71(Supplement_1):532-P. doi.org/10.2337/db22-532-P
Deja S. et al. Diabetes 2022;71(Supplement_1):532-P. doi.org/10.2337/db22-532-P In vivo infusion of stable isotope tracers through jugular vein catheter enables investigation of metabolism in conscious and unrestrained animals.
A typical protocol is presented in the schematic on the left, where following an overnight fast, stable isotope infusions of [U-13C3]propionate, [3,4-13C2]glucose and 2H2O were performed in conscious and unrestrained animals.
Simultaneous administration of multiple tracers allow for comprehensive labeling of hepatic metabolism which can be evaluated by analyzing blood glucose. A 2H2O bolus provided source specific labeling of newly produced glucose. [U13C3]propionate infusion labels TCA cycle intermediates and allows estimation of anaplerosis and TCA cycle turnover. Finally, [3,4-13C2]glucose is used to measure glucose turnover and to convert relative fluxes to absolute fluxes.
Position specific fragments of glucose provide additional information for the model, allowing deconvolution of 2H and 13C isotopomer labeling patterns and flux estimation based on an iterative procedure of error minimization.
Model can be expanded for organic acids and amino acids to provide tissue specific additional information for the model, allowing determination additional fluxes.
Join Our Lab
Be part of the great impact we're having on science and medical care across the globe.We are actively looking for graduate students and postdoctoral researcher candidates.