Positioning of mitochondrial cristae and functions of the MICOS complex
Cristae membranes are the hallmark feature of mitochondria, giving them their distinctive appearance in electron microscopy images. Cristae are home to the respiratory complexes that generate ATP, which makes us think of mitochondria as the "powerhouse of the cell" (among their multitude of other jobs!).
Despite it being such a fundamental question how cristae are made, there is still a lot we don't understand. The key protein complex that contributes to cristae organization, MICOS, was only discovered about 10-15 years ago. MICOS is highly conserved and plays a crucial role in stabilizing cristae junctions, the sites of cristae invaginations. However, we think that MICOS likely plays additional roles through its interactions inside of mitochondria.
We are interested in the mechanisms that control MICOS positioning within mitochondria and how MICOS senses and responds to cellular metabolic needs to dynamically organize cristae membranes and mitochondrial functions. We recently found that a key factor in contributing to cristae organization comes from outside the organelle, as we found MICOS assemblies are positioned near sites where mitochondria contact the endoplasmic reticulum. Check out this work here!
 An electron micrograph of a mitochondrion in human fibroblasts
An electron micrograph of a mitochondrion in human fibroblasts Regulation of selective mitochondrial turnover by autophagy
There are many different pathways responsible for ensuring mitochondrial health. However, in a number of different circumstances, mitochondria need to be destroyed. This can occur developmentally as some cell types like red blood cells need to remove all their mitochondria. However, individual mitochondria or portions of mitochondria can be removed during acute stresses or even for housekeeping purposes through the selective autophagy of mitochondria (mitophagy).
Recently, we discovered an outer mitochondrial membrane protein, TMEM11, that interacts with both the MICOS complex and proteins responsible for one type of mitophagy. We think that TMEM11 may help decide where mitophagy is triggered within the mitochondrial network. Currently, we are exploring the possibility that MICOS communicates internal mitochondrial dysfunction to the mitophagy machinery through TMEM11.
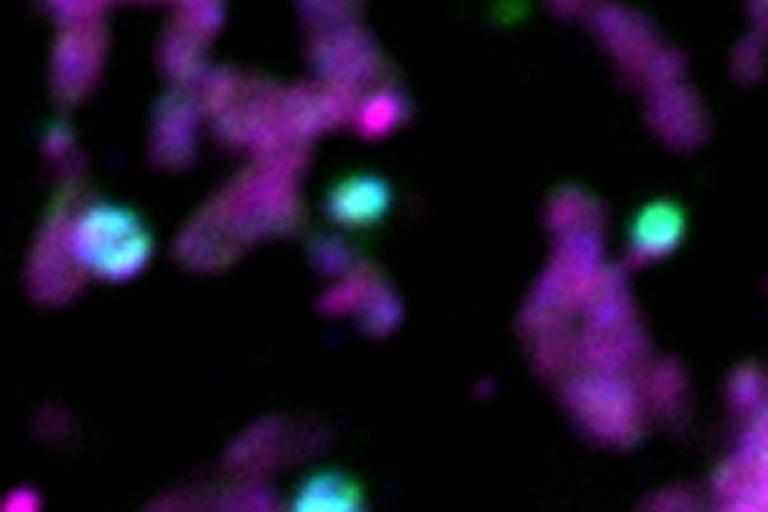 Sites of mitophagy (marked in blue and green) of portions of the mitochondrial network (magenta) in human cells
Sites of mitophagy (marked in blue and green) of portions of the mitochondrial network (magenta) in human cells Coordination of mitochondrial inner membrane remodeling with organelle fission
Mitochondria are highly dynamic organelles, moving around along the cellular cytoskeleton and changing their shape and distribution through division and fusion. Mitochondrial fission, the process of dividing a mitochondria into two pieces, is coordinated with contacts between mitochondria and other organelles like the endoplasmic reticulum. Additionally, the mitochondrial genome, located in the mitochondrial matrix, undergoes replication that is positioned at the fission site. This raises a key question of how mitochondrial fission can be coordinated between the outside and inside of the organelle.
Recently, we identified a new player in mitochondria division, a protein that resides inside the organelle that is required for the external machinery to complete fission. This provides a new opportunity to understand how communication occurs across the two membrane bilayers of mitochondria.
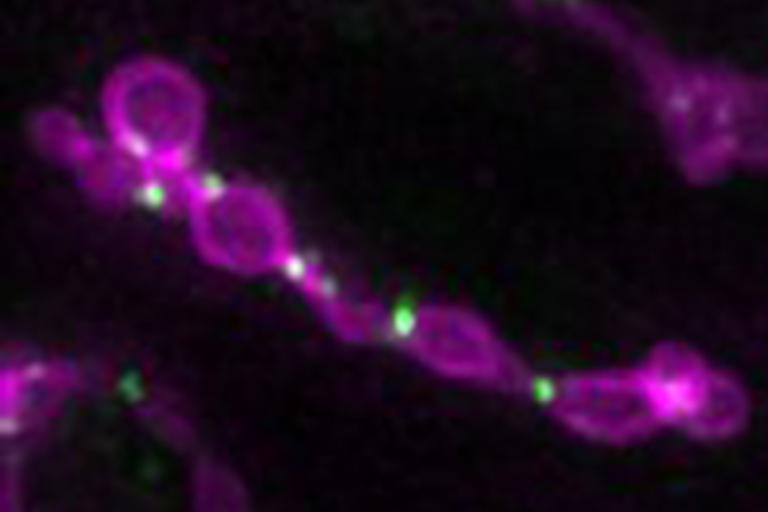 Mitochondrial fission machinery (green) stalled on dividing mitochondria in fission yeast
Mitochondrial fission machinery (green) stalled on dividing mitochondria in fission yeast A key phospholipid synthesizing enzyme cycles between building mitochondria and making fat storage organelles
Mitochondrial phospholipids play a key role in establishing the shape and the respiratory function of the organelle. The mitochondrial inner membrane is the site of synthesis of two key cellular phospholipids, cardiolipin and phosphatidylethanolamine (PE). Remarkably, the mitochondrial PE synthesis enzyme, Psd1, can work in more than one place in the cell and dynamically targets to the endoplasmic reticulum to varying degrees depending on nutrient availability.
Recently, we discovered that the reason for the dual targeting of Psd1 is to provide phospholipids to build a fat storage organelle, the lipid droplet. We are currently working to understand how the cell can coordinate opposing metabolic processes, storage and utilization of fat, through the dynamic targeting of a single enzyme.
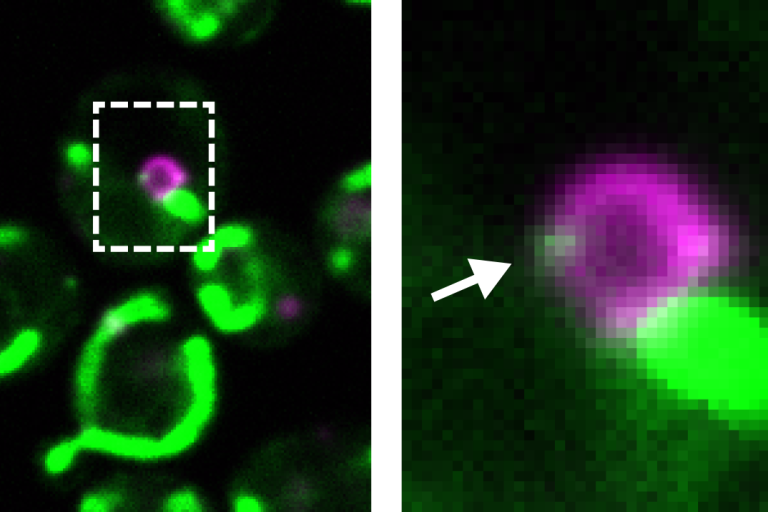 Psd1 (green) simultaneously localizes to mitochondria and to the base of growing lipid droplets (magenta)
Psd1 (green) simultaneously localizes to mitochondria and to the base of growing lipid droplets (magenta)