The transmission of arboviruses, such as the Zika virus, from Dipteran insects (flies and mosquitoes) to mammalian hosts continues to pose significant economic and public health challenges. In light of this, there has been much interest in using Dipteran model organisms to gain insights into arbovirus-host interactions and explore eukaryotic antiviral innate immunity in general.
However, because these arboviruses are well-adapted to Dipterans, it may be difficult experimentally to use these model systems as tools to identify conserved host antiviral defense mechanisms that are already effectively countered by these viruses. Furthermore, studying arboviruses in alternative eukaryotic hosts may lead to the identification of cellular restriction mechanisms that are absent in Dipteran insects.
While classic approaches have focused on examining productive infections of a host cell, we have developed a new paradigm for probing virus-host interactions that exploit naturally abortive infections. Such infections can provide important insights into antiviral immunity that could be masked in settings where viruses have evolved multiple and effective immune evasion strategies.
Our strategy draws from our discovery that several arboviruses undergo abortive infections in cells derived from Lepidopteran insects (moths and butterflies). However, arbovirus replication can be “rescued” when Lepidopteran cells are co-infected with a second virus encoding immunomodulatory factors that suppress the cellular immunity mechanisms impeding arbovirus replication.
The fact that these RNA viruses are unable to overcome Lepidopteran antiviral responses on their own makes them excellent tools for identifying conditions that promote virus replication. Using Lepidopteran cell lines, RNA viruses encoding convenient reporters (i.e. GFP, luciferase), and RNA interference (RNAi) screening, we have developed sensitive reporter-based assays to identify both host factors that restrict and viral “immunomodulators” that promote, arbovirus replication.
These systems are also amenable to the genetic or plasmid-based mapping of candidate viral immunomodulators. Using these screening tools, we have identified, for the first time, several Lepidopteran antiviral pathways that impede RNA virus replication, including pathways that are conserved in mammals.
We are currently characterizing these antiviral pathways further to apply this knowledge to mammalian host systems.
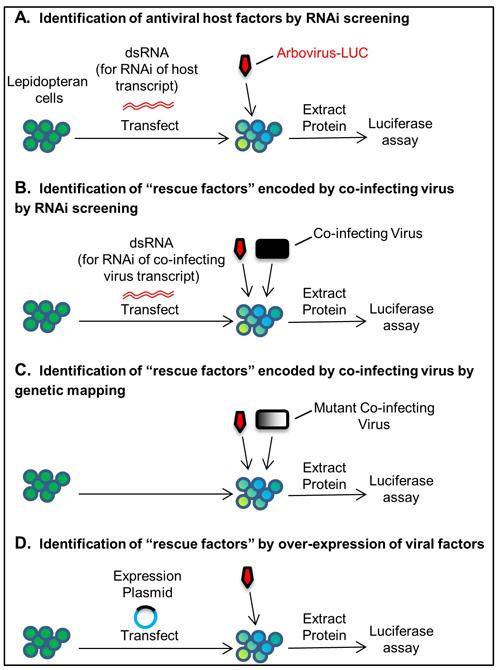 Lepidopteran assay systems for identifying host antiviral factors and virus-encoded factors that “rescue” arbovirus replication. Assays shown employ arboviruses encoding a luciferase reporter gene (Arbovirus-LUC). Luciferase (light) signals from cell lysates serve as readouts of arbovirus replication under each experimental condition.
Lepidopteran assay systems for identifying host antiviral factors and virus-encoded factors that “rescue” arbovirus replication. Assays shown employ arboviruses encoding a luciferase reporter gene (Arbovirus-LUC). Luciferase (light) signals from cell lysates serve as readouts of arbovirus replication under each experimental condition.