In addition to amino acid substitutions, we are interested in exploring other adaptive strategies used by hosts to defend against invading pathogens. We recently hypothesized (Hancks et al. 2015) that protein isoforms generated via splicing may be an additional strategy to counteract pathogen-encoded proteins which directly bind host proteins to modify their function (see model below). Along with testing this model, we are also interested in identifying and characterizing other countermoves used by the host to defend itself.
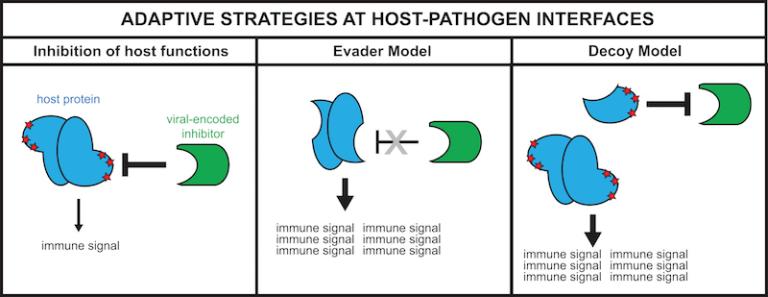 Figure 5
Figure 5 Figure 5 shows host adaptive strategies to evade pathogen-mediated antagonism. A model we generated based on observations made while characterizing the evolution of cGAS (Hancks et al. 2015). Signatures of rapid evolution (red stars) can often reveal protein surfaces targeted by pathogen-encoded inhibitors (green). Along with amino acid substitutions, alternative splicing may be a means to generate protein isoforms that remove protein surfaces targeted by the inhibitor (evader model) and/or generate decoy proteins that function as competitive inhibitors (decoy model).