Pathogens must manipulate cellular functions to replicate and evade host defenses. The ability of the host (cell) to counteract such antagonism directly impacts the outcome of infection. A major goal of our research is to identify these novel tipping points of infection. The usual suspects include host proteins that function as first responders to infection (e.g. pattern-recognition receptors, sensors) with well-known examples being protein kinase R (PKR) and other interferon-stimulated genes (ISGs). Alternatively, host factors that control processes such as nucleic-acid metabolism are likely to represent pivotal interfaces during infection due to the requirement of a pathogen to replicate its genome as part of its lifecycle.
Pathogens commonly manipulate host functions by directly binding cellular proteins. Hosts can “escape” this attack by a pathogen via a cycle of mutation and selection. Specifically, as mutations occur, mutations are selected for on the host side which destroys this interaction. These dynamics resemble an “arms race” at the molecular level and are a type of genetic conflict. These predator-prey interactions were first predicted by Leigh Van Valen’s “Red Queen Hypothesis”.
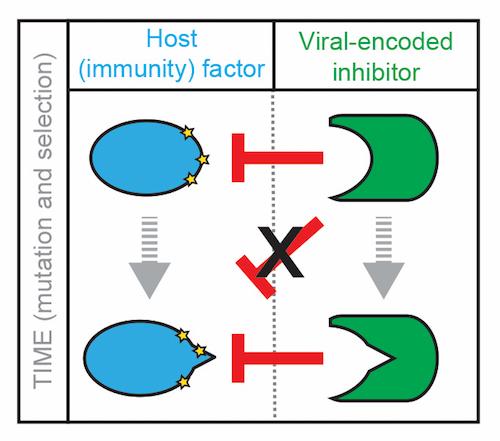 Figure 1
Figure 1 Figure 1 shows a model of host-pathogen interactions. A host factor (blue), which can determine the outcome of infection by triggering immune defenses or limiting infection by other means, is targeted for inactivation by a pathogen-encoded protein, in this instance a viral protein (green). Inhibition of the host protein is detrimental to host fitness such that mutations are selected which weaken this interaction. Notably, mutations also occur on the viral side such that mutations that restore this interaction are selected. Using sequence information and evolutionary analysis we can gain insight into this “molecular tug-of-war”. Yellow stars represent amino acid positions that are rapidly evolving. The major goal of this project is to identify such antagonistic relationships and how the biology of the host and pathogen proteins impact infection outcomes.
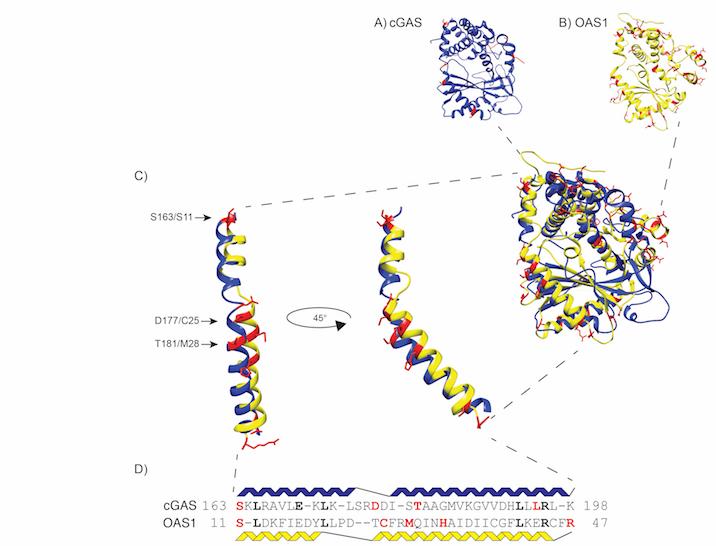 Figure 2
Figure 2 Figure 2 shows evolutionary analysis can reveal novel patterns of pathogen-mediated antagonism. Our work on the evolutionary history of two major antiviral proteins which sense foreign nucleic acids – cGAS and OAS1 – uncovered a common protein surface likely targeted by distinct pathogens to facilitate the inactivation of these proteins. Interestingly, a subset of the signatures of positive selection between the two proteins was shared. These data perhaps hint at a yet undiscovered pathogen-encoded protein that can bind and inhibit both cGAS and OAS1 (Hancks et al. 2015). Rapidly evolving amino acid positions are highlighted in red.