Regulation and function of microRNAs
miRNAs are an abundant and diverse family of ~20- to 23-nucleotide RNAs that recognize sites of complementarity in target mRNAs, resulting in decay and reduced translation of target transcripts. Approximately 500-1000 miRNAs are encoded in mammalian genomes. Our work on the miRNA pathway focuses mainly on three broad questions: What functions do miRNAs perform in normal physiologic states? How does aberrant miRNA activity contribute to diseases such as cancer? How is miRNA abundance and activity regulated in normal physiology and disease?
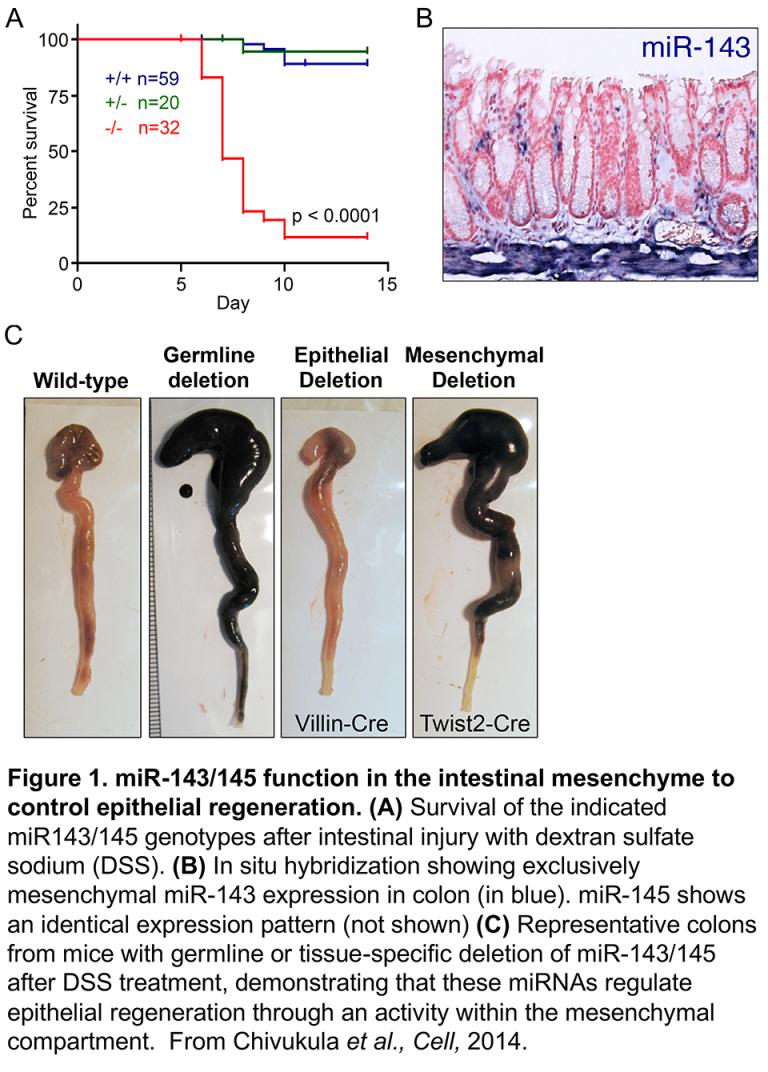
In addressing these questions, we emphasize the use of in vivo models to robustly dissect miRNA function. The importance of studying miRNAs in vivo is exemplified by our analysis of the miR-143/145 cluster (Chivukula et al., Cell, 2014), one of the most widely studied anti-tumorigenic miRNA loci. Downregulation of miR-143/145 in colorectal tumors was one of the first reported miRNA abnormalities in human cancer and over 200 subsequent papers implicated these miRNAs as epithelial-intrinsic tumor suppressors. Nevertheless, the natural functions of these miRNAs in epithelial tissues had not been studied. We generated miR-143/145 conditional knockout mice to address this question. Our studies of these animals unexpectedly revealed that this miRNA cluster is exclusively expressed in the mesenchymal compartment of intestine, where it performs an essential function in promoting epithelial repair following injury (Figure 1). These findings provided important new insights into mechanisms of injury repair and clarified the role of this miRNA cluster in cancer.
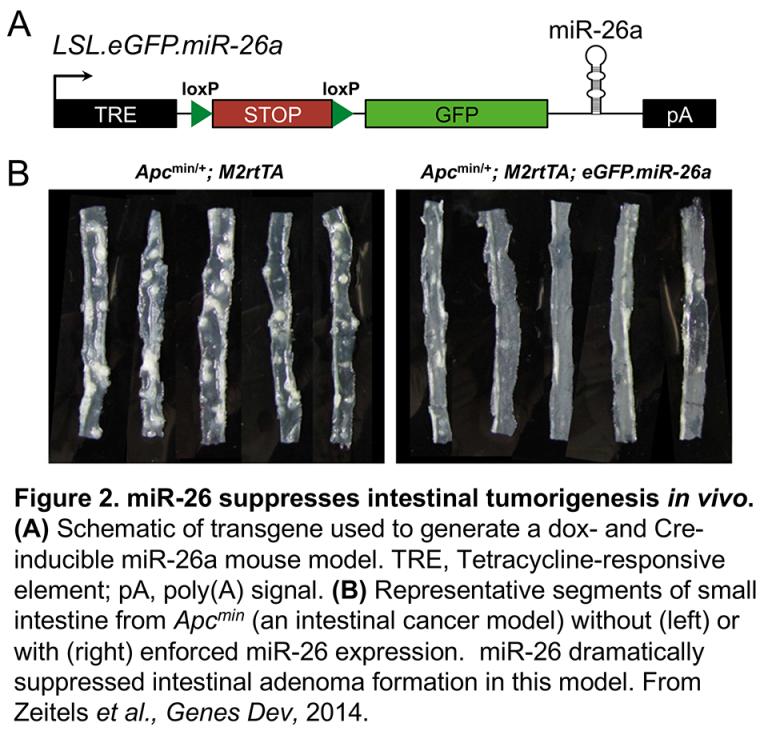
Our laboratory has studied other important miRNAs using in vivo models, including miR-122 (Hsu et al., JCI, 2012) and the miR-26 family (Zeitels et al., Genes Dev, 2014) (Figure 2). CRISPR-mediated genome editing has dramatically enhanced our ability to generate gain- and loss-of-function models to study miRNA biology, representing an important aspect of our ongoing research program.