Benign genetic variation
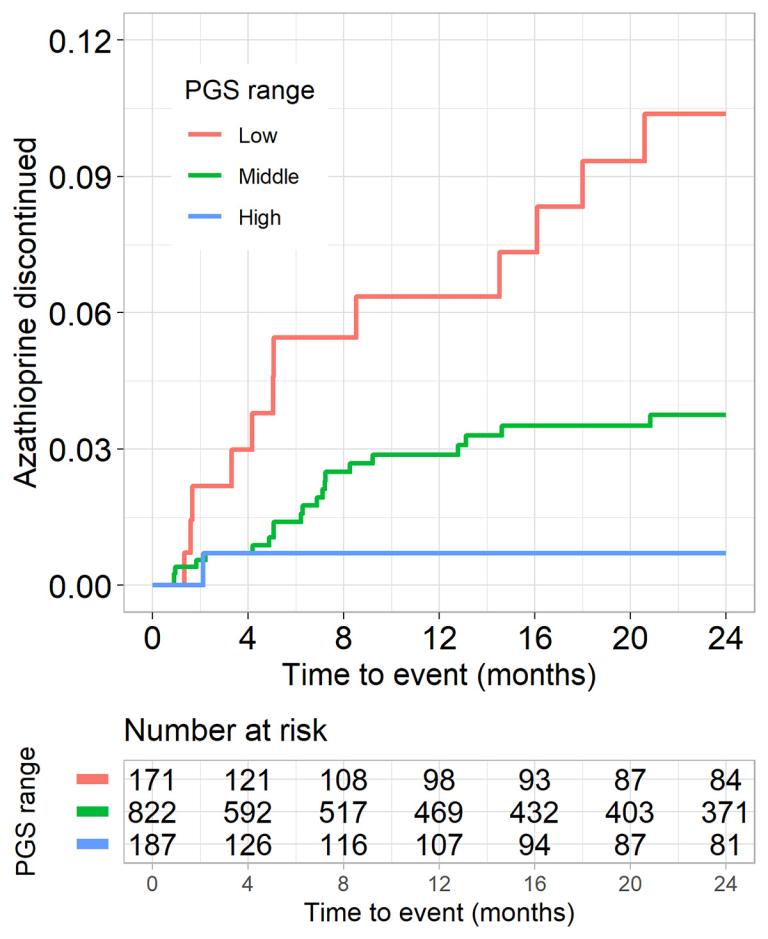
Dynamic biomarkers are frequently measured in clinical settings to screen for underlying disease and guide care. Longitudinal studies have demonstrated that a healthy individual’s biomarker levels tends to fluctuate around a distinct set points and, for some individuals, their set point lies near or exceeds clinical decision thresholds, which predisposes them to alterations in care in the absence of disease. The levels of dynamic biomarkers within a population are heritable, with an important contribution from polygenic variation, which can be measured. While this genetic variation is intrinsically “benign”, we have demonstrated it leads to vulnerable populations predisposed to iatrogenic harms (such as invasive diagnostic testing, inappropriate cessation of medications, and prolonged diagnostic odysseys) due to clinical providers misinterpreting the reason for an outlying value. One focus of the lab is improving clinical decision-making with respect to benign variation with the goals of guiding clinical decisions around therapeutics, shortening or avoiding diagnostic odysseys by providing an explanation for outlying biomarker levels, and ensuring screening strategies do not systematically create populations vulnerable to under- or over-diagnosis.
Biomarker discovery and characterization
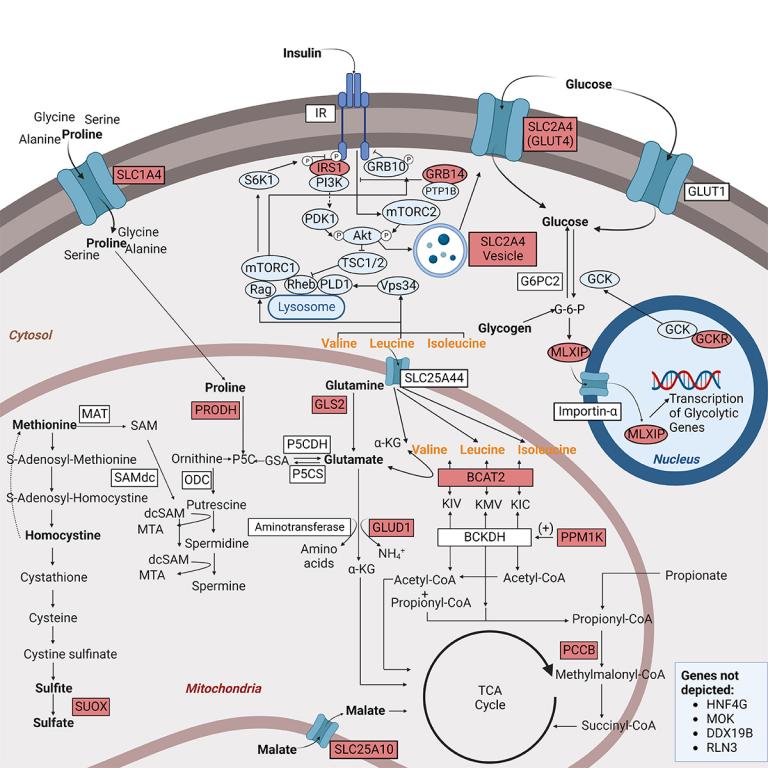
Biomarkers are reproducible measures of a physiological state and can facilitate early diagnosis and risk stratification and, in instances where the biomarker is a mediator of disease, can be targeted to prevent or treat disease. One focus of the lab is to leverage polygenic approaches to define the etiological relationships between biomarkers, such as metabolomic and proteomic markers, and disease. Variation in many biomarkers is polygenic, and an advantage of understanding variability in biomarker levels from a genetic context is that discrete mechanisms contributing to variability can be individually interrogated to understand their independent associations with outcomes. We have implemented discovery-oriented phenome wide association (PheWAS) approaches to screen for biomarkers associated with clinically derived phenotypes. We also use hypothesis-testing approaches such as Mendelian Randomization to define the basis of epidemiological associations between biomarkers and disease and have shown, for instance, that the association between thyroid stimulating hormone risk and atrial fibrillation risk is mediated, in part, by growth effects of thyroid hormones; and the branched chain amino acids (BCAAs) are likely early biomarkers of type 2 diabetes and not mediators of risk.
Polygenic risk prediction of disease risk
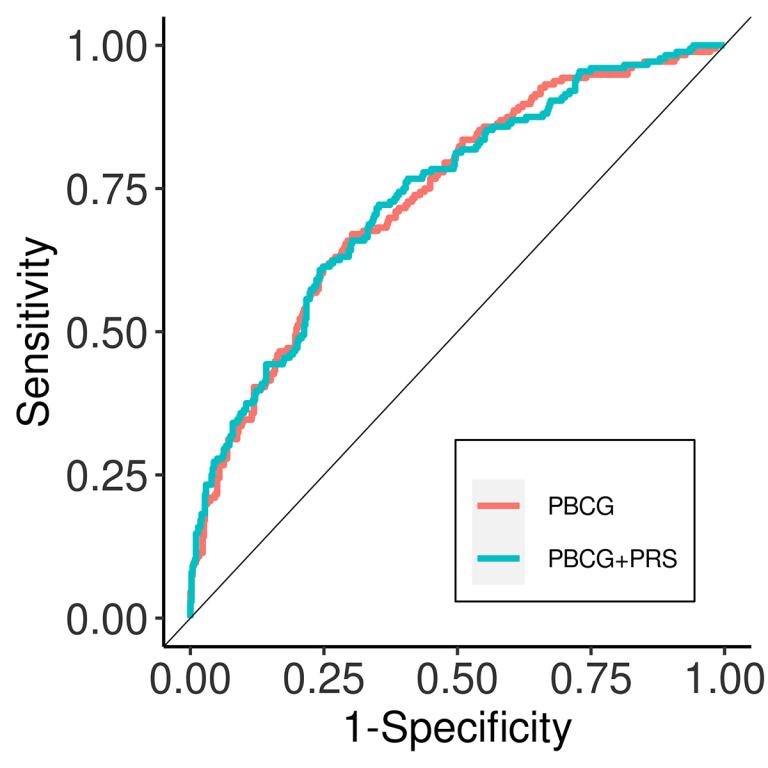
Risk for many complex, chronic diseases, such as cardiovascular diseases, are influenced by polygenic variation. Consistently, genome-wide association studies have identified large numbers of SNPs associated with chronic diseases and risk factors. There is considerable interest in applying polygenic predictors of clinical disease to clinical practice. In evaluating predictors, it is essential that they be benchmarked against clinical practice standards and clinically relevant outcomes. We showed, for instance, that a polygenic risk score of coronary artery disease only modestly improved risk prediction over standard clinical tools, and that polygenic predictors of prostate cancer risk did not improve discrimination for low grade cancers versus clinically significant high grade cancers. One focus of the lab is identifying clinical settings and genetic tools that improve clinical risk stratification.