Our research aims to obtain a comprehensive picture of how genomic stability and chromatin dynamics affect neuronal functions, including learning behaviors, and to apply this knowledge to combat neurological disorders.
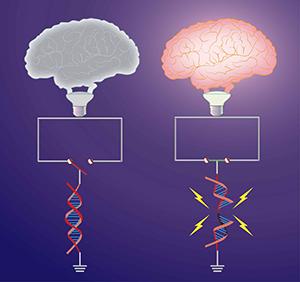
Damage to the genetic material of cells is thought to be a major cause of tissue dysfunction. The nervous system is particularly vulnerable to genomic instability, and mutations in DNA damage response factors manifest with severe neurological abnormalities. Yet, precisely how DNA damage contributes to the development of neurological disorders remains an important unresolved question, and the nature of lesions and the sources of DNA damage that are most pertinent to neuronal dysfunction remain unknown. In this regard, we and others recently reported that numerous patterns of physiological neuronal activity, including learning behaviors, are associated with the formation of DNA double strand breaks (DSBs). Our studies on these activity-induced DSBs have led to the following findings:
- Activity-induced DSBs form at highly specific loci within the neuronal genome. Surprisingly, these loci are enriched for the promoters of a subset of immediate early genes (IEGs), including Fos, Npas4, FosB, Nr4a1, and Egr1, that play crucial roles in experience-driven synaptic changes, learning and memory
- Activity-induced DSBs are generated through the actions of a topoisomerase, topoisomerase IIb (Top2B)
- These Top2B-mediated DSBs facilitate the rapid induction of the aforementioned IEGs following appropriate neuronal stimulation
The unexpected link between DSB formation and neuronal activity-dependent gene expression suggests that changes in the ability to either form or accurately repair activity-induced DSBs could impact cognitive performance and lead to the development of neurological disorders. Our research efforts therefore focus on elaborating the molecular mechanisms that regulate the formation and repair of activity-induced DSBs in neurons.
Furthermore, it is now clear that the relationship between Top2B-mediated DSBs and stimulus-dependent transcription is not unique to neurons, but is also seen in genes activated by serum, estrogen, androgen, glucocorticoid, and insulin signaling. An additional interest of the lab is to understand whether aberrant repair of Top2B-mediated DSBs in these systems could have pathological implications, such as the development of cancer.
 Madabhushi Lab Research
Madabhushi Lab Research