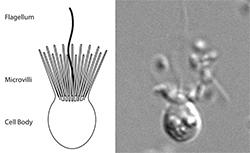
Host-pathogen interactions are widespread in nature, and all organisms, from bacteria to mammals, likely have pathogen response systems. However, how animal innate immune responses first evolved is unclear. While major discoveries in immunity have come from studies in mammals, examining infection and host response in other diverse organisms has illuminated important pathways ranging from Toll to RNAi to CRISPR. The complexity and redundancy of mammalian immune response pathways may mask novel sensors and signaling networks, limiting their discovery and exploitation. Thus, we are examining infection and host response in choanoflagellates, the closest living relatives of animals. Choanoflagellates are aquatic single-celled and colony-forming eukaryotes that have diverse obligate interactions with bacteria; not only do choanoflagellates feed on bacteria, but specific bacterial molecules induce choanoflagellate multicellularity and mating. Therefore, choanoflagellates can illuminate animal origins and specific mechanisms by which bacteria influence eukaryotic biology. However, it remains unclear how choanoflagellates defend against infection. Genomes and/or transcriptomes from 18 choanoflagellate species provide tantalizing hints about which known immune pathways may be present in choanoflagellates. Choanoflagellates lack evidence for many canonical innate immune pathways, for both bacterial and viral sensing and response. For example, choanoflagellates lack genes required for most known bacterial sensing pathways, and they lack downstream effectors such as death kinases. Similarly, choanoflagellates do not have the genes required for RNAi in other eukaryotes as well as known pathways that sense double-stranded RNA, a hallmark of viral infection. Do viruses infect choanoflagellates, and if so, are they sensed? Although choanoflagellates feed indiscriminately on bacteria, how do they respond to pathogenic bacteria? We are identifying and characterizing innate immune factors in choanoflagellates using both unbiased and hypothesis-driven approaches. Overall, this work has the potential to identify new innate immune sensing mechanisms, some of which may be conserved in animals. Examining innate immunity in this completely untapped system has the potential to reveal unique biology in the same way that studies in flies led to the discovery of Toll and studies in worms led to the discovery of RNAi.
In other recent work, we have been examining the impact of circadian rhythms on enteric virus infection. Circadian rhythms are 24 hour oscillations in a variety of processes that are entrained by environmental cues including light. Molecularly, this “clock” is driven by key transcription factors and feedback loops that generate rhythmic expression of thousands of mammalian genes in a variety of tissues. Past work has revealed the impact of circadian rhythms on metabolism and immunity. However, the impact of circadian rhythms on infection, particularly enteric virus infection, is understudied. Preliminary experiments using coxsackievirus B3 revealed a profound circadian effect on infection. Our ongoing work is examining the mechanisms and consequences of host rhythmic gene expression on enteric virus infection.