The Phillips Lab studies the function and regulation of biochemical pathways in protozoal parasites with the goal to exploit these pathways for new drug discovery. Major projects include:
- The polyamine biosynthetic pathway in Trypanosoma brucei
- Drug discovery projects targeting the malaria parasite Plasmodium falciparum including: 1) targeting the pyrimidine biosynthetic pathway, and in particular the enzyme dihydroorotate dehydrogenase, 2) targeting the proteasome, and 3) identifying new potential targets of anti-malarial compounds identified in a phenotypic screen.
Malaria
The human malaria parasite is endemic in 87 countries, putting 2.5 billion people in the poorest nations of the tropics at risk for the disease. Despite intensive efforts to control malaria through combination drug therapy and insect control programs, malaria remains one of the largest global health problems. Widespread drug resistance has compromised the effectiveness of malaria control programs.
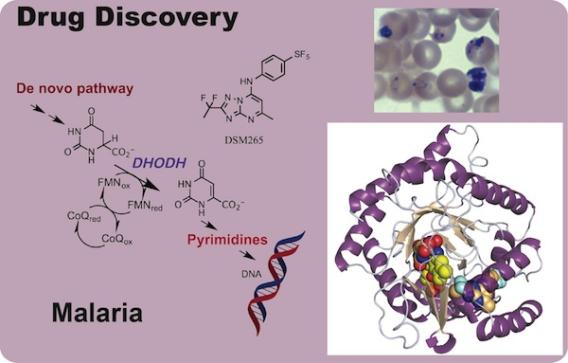
Pyrimidine biosynthesis provides a significant opportunity for the development of new chemotherapeutic agents against the malaria parasite. Plasmodium species rely exclusively on de novo pyrimidine biosynthesis to provide precursors for DNA and RNA synthesis. Blocking pyrimidine biosynthesis would selectively kill the parasite; mammalian cells would be resistant because they have salvage pathways to reuse DNA and RNA precursors.
Our research focuses on dihydroorotate dehydrogenase (DHODH), which catalyzes the fourth step in de novo pyrimidine biosynthesis. We identified a number of chemical scaffolds that are potent and species selective inhibitors of P. falciaprum DHODH by high throughput screening. A triazolopyrimidine-based series emerged from this screen that has potent activity against malaria parasites in vitro, and which suppress parasites in a mouse model of the disease. We solved the X-ray structures of PfDHODH bound to inhibitors from this series utilizing the X-ray structure to inform the medicinal chemistry to optimize both the potency and in vivo pharmacokinetic properties of the compound series. We successfully identified a clinical candidate from this work (DSM265), that reached Phase II clinical trials. Recently we undertook a lead optimization program around a pyrazole-based series that resulted in the discovery of several late-lead candidates with improved properties over DSM265. This effort is part of a multidisciplinary project that is funded by the NIH and by Medicines for Malaria Venture (MMV) to develop our lead compounds into new anti-malarial chemotherapy. The team includes the Phillips Lab, Susan Charman’s group at Monash University (ADME), and a collaboration with Schrödinger (computational drug design). We also have an ongoing project to use X-ray structure and biophysical methods to further understand drug resistance risk against the DHODH target.
New directions in malaria research in the lab extend to identifying new targets for drug discovery and exploiting the proteasome as a target in collaboration with the Ready Lab at UT Southwestern, and with MMV.
Sleeping Sickness
Human African trypanosomiasis (HAT) is caused by the parasitic protozoan, Trypanosoma brucei. Fifty million people are at risk for the disease, which is fatal if not treated. There is a great need to translate recent advances in the understanding of the basic biology of the parasite into new drugs.
Polyamine biosynthesis in T. brucei
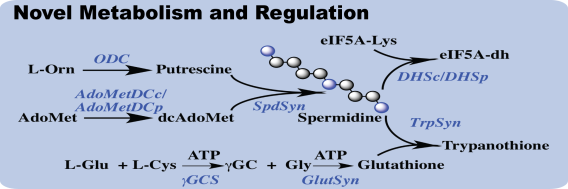
Polyamines are small organic cations that are required for cell growth and in the kinetoplastid parasites and the metabolic pathway is novel in the parasites. The polyamine spermidine is used to form a novel conjugate with glutathione (trypanothione) and the mechanism of polyamine regulation differs from other eukaryotic cells. The pathway has proven to be a rich source for the identification of anti-trypanosomal compounds and the biosynthetic enzyme ornithine decarboxylase (ODC) is the target of eflornithine, the only clinically used compound with a known mechanism of action.
Our lab goals are to understand the function of polyamines in the parasite, to learn how the parasite regulates the pathway and to identify additional targets to be exploited for the development of new drugs. We systematically explored the structure and function of key enzymes in the pathway including ODC, S-adenosylmethionine decarboxylase (AdoMetDC), and more recently, deoxyhypusine synthase (DHS), which conjugates spermidine to the essential translation factor eIF5A to form the hypusine modification that is required for its activity.
We use genetic approaches and metabolite profiling to identify essential enzymes and to learn how the pathway is regulated. Key recent findings include our discovery that both AdoMetDC and DHS are activated by formation of multimers with inactive paralogs – which we termed prozymes – by mechanisms that are specific to parasitic protozoa. We are currently focused on understanding this regulation for DHS at a molecular level through CryoEM structural