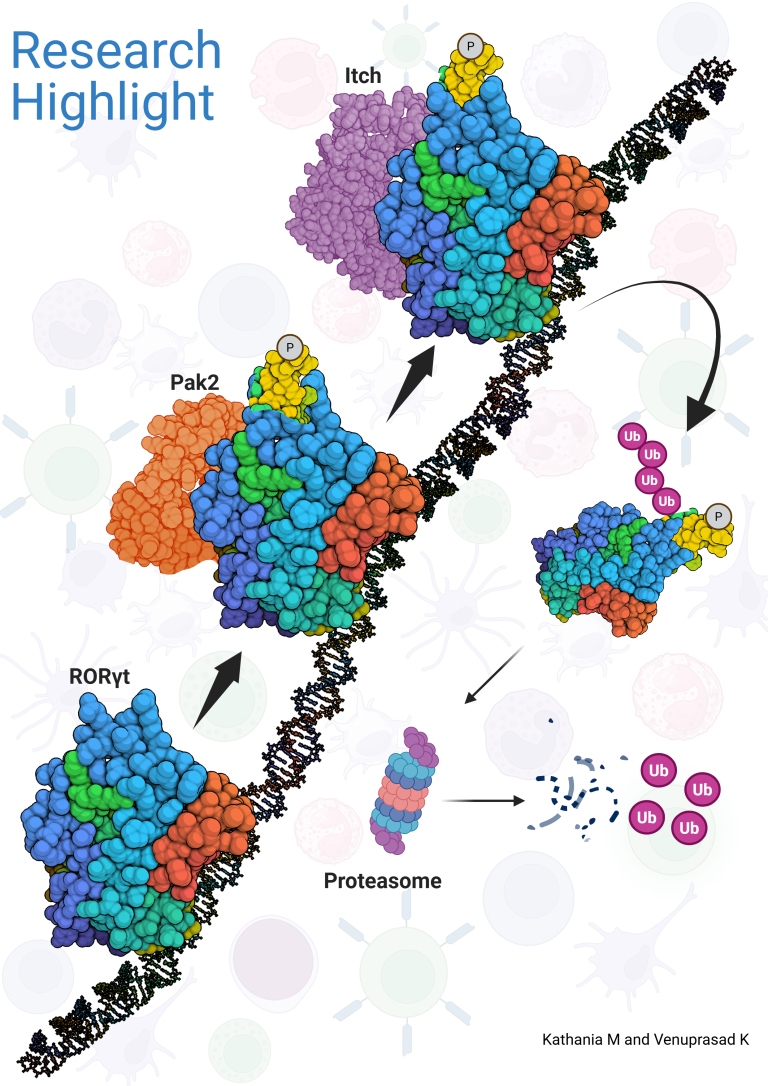
Research
Deep learning for cardiopulmonary MRI processing
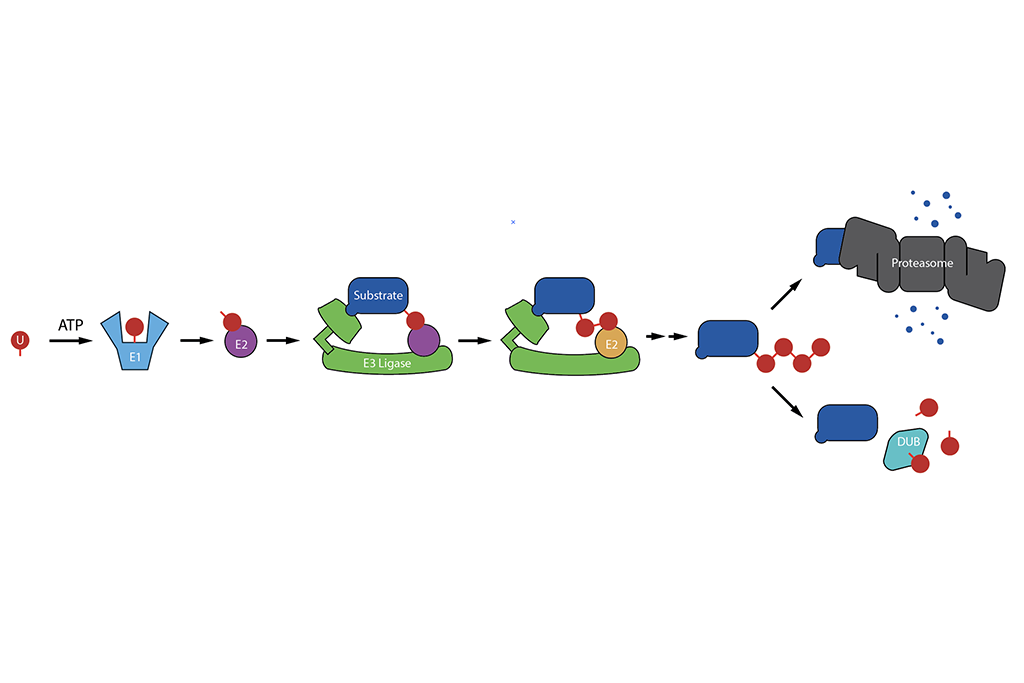
One of the research objects in the lab is on the development of deep-learning algorithms for cardiopulmonary MRI processing, especially the development of unsupervised learning algorithms. We are interested in 2D and 3D cardiopulmonary MRI reconstruction, denoising, super-resolution and segmentation. In this objective, we have developed several algorithms such as G-SToRM, MoCo-SToRM, V-SToRM for real-time cardiopulmonary processing. These work on real-time cardiopulmonary processing wothe best paper award in 2020, and the best paper award finalist in 2021 at IEEE International Symposium on Biomedical Imaging (IEEE ISBI). We've also extended the above algorithms for better performances based on recent advances in deep learning such as the inclusion of Spatial Transformer Networks (STN), Implicit Neural Representations (INR), Graph Convolutional Network (GCN),Kernel method into the algorithms.
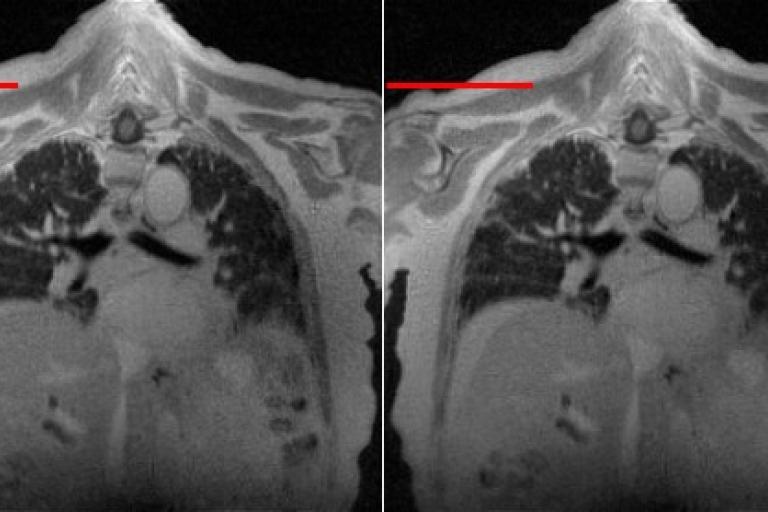 3D Lung MRI
3D Lung MRI 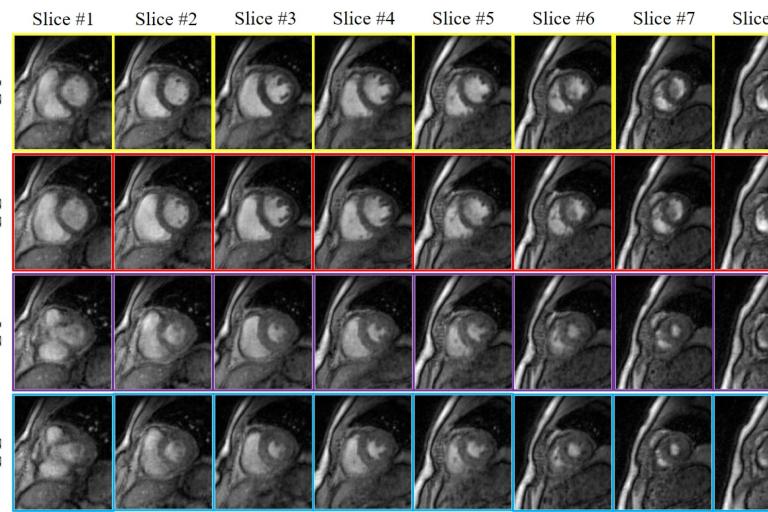 Multi-slice Cardiac Cine
Multi-slice Cardiac Cine 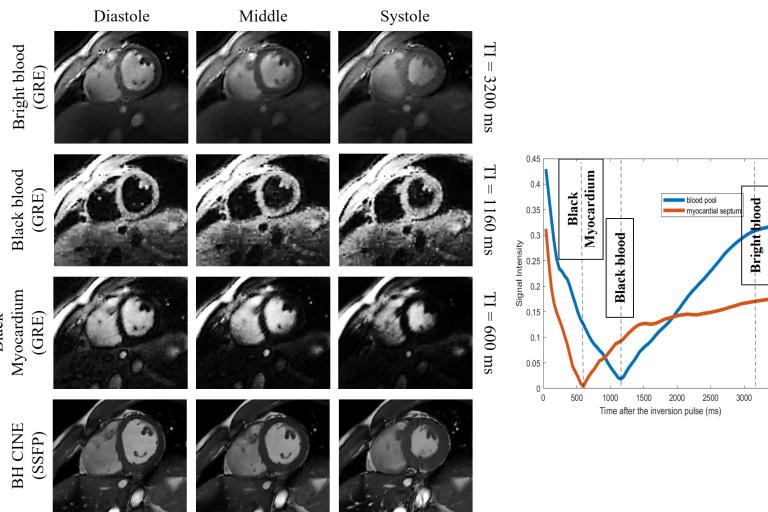 Multi-Contrast cardiac Cine
Multi-Contrast cardiac Cine Pulse sequences development for cardiopulmonary MRI
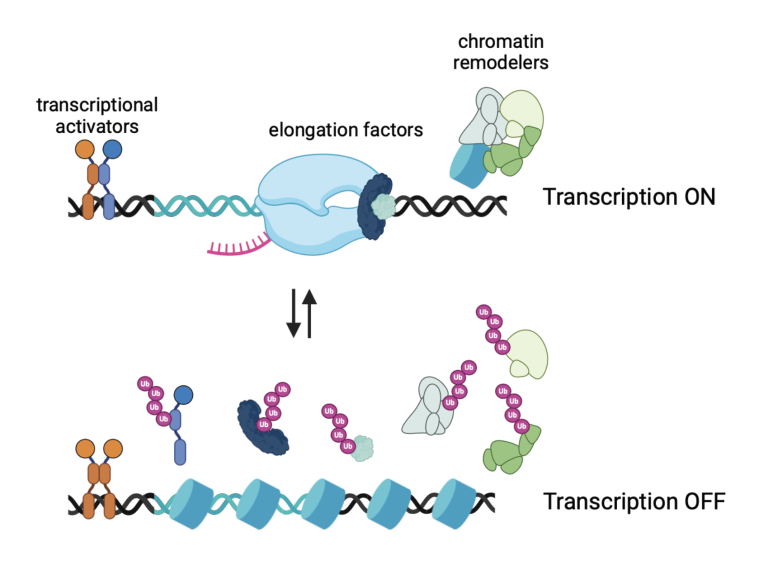
The second line of research in the lab is on the development of novel fast 2D and 3D cardiopulmonary MRI pulse sequences for the care of patients with congenital and acquired heart diseases. This includes the development of fast 2D or 3D sequences for myocardial quantification, such as multi-tasking sequences, MR fingerprinting sequences. We are also interested in developing robust fast 3D MR angiography sequences, 3D lung sequences based on ultra-short echo time (UTE) sequences and non-contrast-enhanced 3D whole-heart sequences
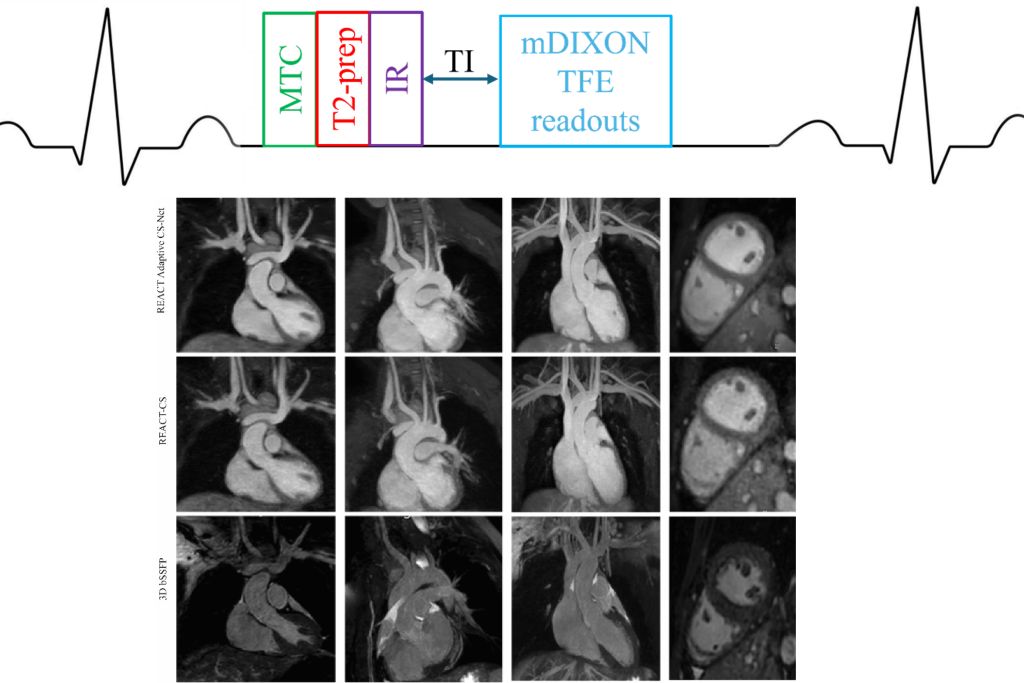 Non-Contrast-enhanced 3D whole-heart MRI
Non-Contrast-enhanced 3D whole-heart MRI Translational cardiology research using advanced Cardiac imaging
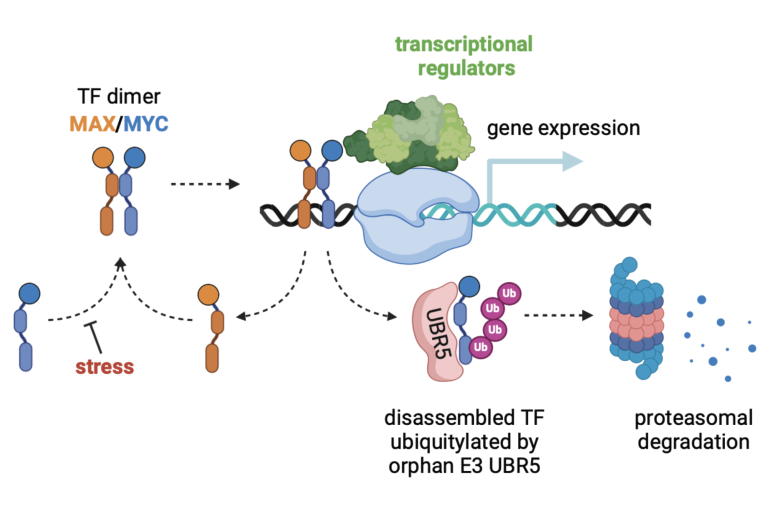
Our lab also shows interest in developing new imaging biomarkers for congenital and acquired heart diseases. For example, we would like to develop new image biomarkers based on Magnetization transfer (MT) and Chemical exchange saturation transfer (CEST) for the quantification of myocardium without using Gadolinium-based contrast agents (GBCAs) for patients with acquired heart diseases. We are also interesting in developing new quantitative imaging biomarkers for the monitor of diseases progression for patients with congenital heart diseases, especially the patients with single-ventricle physiology.
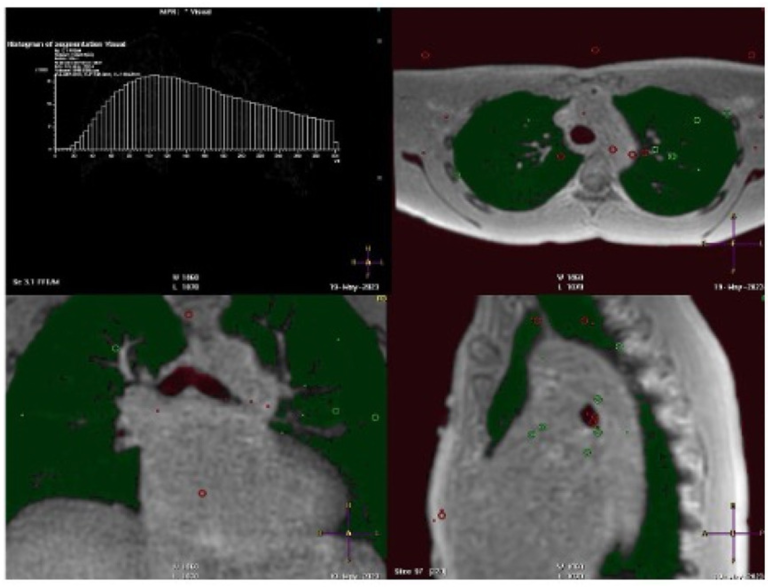 Quantify the level of water in the lung
Quantify the level of water in the lung Research projects at Zou Lab at UT Southwestern