Discovery of New Drugs for the Treatment of Lung Cancer
The objective of this Cancer Prevention and Research Institute of Texas (CPRIT) project is to identify and target lung cancer acquired vulnerabilities (“synthetic lethalities”) that have arisen during lung cancer pathogenesis and that can be used as new, cancer specific therapeutic targets with associated molecular “enrollment biomarkers” for personalized treatment.
When oncogenic changes (mutations and epigenetic changes) arise during lung cancer development they represent absolute differences from normal tissues that have the potential to become lung cancer specific therapeutic targets. While notable examples of this are acquired mutations in oncogenes such as the EGFR receptor and its targeting by EGFR tyrosine kinase inhibitors, these actually represent only a minority of cases and targeting these by themselves are not curative.
By contrast, our genome-wide knockout (RNAi) screens, as well as our large compound library screens (220,000 synthetic chemicals and natural product extracts) have identified “hits” that kill lung cancer but not normal lung epithelial cells and have defined associated molecular alterations which provide ways to identify specifically which patients would respond to such new synthetic lethality targeted therapy.
These studies are done as a collaboration with an important group of UT Southwestern investigators which include Dr. Michael White, Ph.D., (Department of Cell Biology) Drs. Stephen McKnight, Michael Roth, John MacMillan, Joseph Ready, Bruce Posner, Noelle Williams (Department of Biochemistry), and Yang Xie (Department of Clinical Sciences).
In addition we are conducting whole genome sequencing of our large panel of lung cancer cell lines and xenografts (~300), in collaboration with Richard Gibbs, Ph.D., and his team at the Baylor College of Medicine Human Genome Sequencing Center.
These “oncogenotypes” together with our findings regarding selective RNAi and chemical toxin sensitivity in our panel of NSCLC cell lines will eventually make it possible to use whole genome sequencing to genotype biopsy samples of lung cancer, establish the appropriately categorized oncogenotype, then predict the use of the properly tailored therapeutic for each cancer patient.
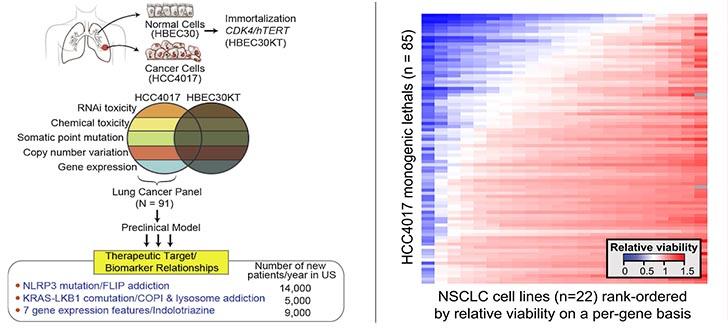
HCC4017, which was derived from a 62-year old female smoker with stage 1A lung adenocarcinoma, was instrumental in discovering three distinct target/response-indicator pairings that are represented with significant frequencies (6%–16%) in the NSCLC patient population (Kim et al, Systematic Identification of Molecular Subtype-Selective Vulnerabilities in Non-Small-Cell Lung Cancer. Cell 155:552, 2013).