Discovery of New Therapeutic Targets using Loss of Function In Vivo screening in Lung Cancer
Advances in “functional genomics” including use of targeted shRNA and CRISPR libraries provide an important opportunity to identify potential therapeutic vulnerabilities in lung cancer that can then be used as a “roadmap” to rationally develop small molecule therapeutics and also assess therapeutic windows. Of course, DepMap and related functional genomic databases have done this in a genome wide fashion. However, all of these have been in vitro screens with tumor cells growing in 2D tissue culture. Thus, we are focusing on targeted screens that are assessed in xenografted tumors growing in vivo – which more closely mimics the clinical situation in patients. By performing simultaneous screens in vivo and in vitro we can determine whether there are major differences in these two approaches. In addition, by using targeted libraries we can both use many guides per gene providing validation of any one target, and also achieve large “representations” of cells tested for each genomic targeting (that is be able to have thousands of tumor cells tested for each guide in vivo to avoid stochastic drop outs. This approach has demonstrated there are dramatic functional differences between what happens in tissue culture and what happens in tumors indicating that we need to rethink our functional genomic dependencies that are only based on tissue culture assays. Here we present two current examples of such approaches we have used in both cases focusing on potentially “druggable” targets.
Functional Genomics of Nuclear Receptors and their Co-Regulators in NSCLC
Using a mini-library of 1,062 lentiviral shRNAs targeting 40 nuclear hormone receptors and 70 of their co-regulators, we searched for potential therapeutic targets that would be important during in vivo tumor growth using a parallel in vitro and in vivo shRNA screening strategy in the non-small cell lung cancer (NSCLC) line NCI-H1819. We identified 21 genes essential for in vitro growth, and nine genes specifically required for tumor survival in vivo, but not in vitro: NCOR2, FOXA1, HDAC1, RXRA, RORB, RARB, MTA2, ETV4, and NR1H2. We focused on FOXA1, since it lies within the most frequently amplified genomic region in lung adenocarcinomas. We found that 14q-amplification in NSCLC cell lines was a biomarker for FOXA1 dependency for both in vivo xenograft growth and colony formation, but not mass culture growth in vitro. FOXA1 knockdown identified genes involved in electron transport to be among the most differentially regulated, indicating FOXA1 loss may lead to a decrease in cellular respiration. In support of this, FOXA1 amplification was correlated with increased sensitivity to the complex I inhibitor phenformin. Integrative ChipSeq analyses reveal that FOXA1 functions in this genetic context may be at least partially independent of NKX2-1. Our findings are consistent with a neomorphic function for amplified FOXA1, driving an oncogenic transcriptional program. These data provide new insight into the functional consequences of FOXA1 amplification in lung adenocarcinomas and identify new transcriptional networks for exploration of therapeutic vulnerabilities in this patient population (PMC7281309).
Functional Genomic Screening Identifies Targets that Enhance Response to Clinically Available Chemotherapy
Taxanes are commonly used as part of platin doublet chemotherapy in the treatment of NSCLC. However, tumor shrinkage and clinical benefit is modest. Thus, we wanted to discover if there were other “druggable” targets that could dramatically sensitize NSCLC tumor cells to taxanes. To achieve this, we collaborated with the laboratory of UTSW scientist Joshua Mendell to perform CRISPR-Cas9 screens. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-directed Cas9-mediated endonuclease activity can disrupt specific genetic sequences in the genome and provide promising tools to perform Loss of Function (LOF) screens. Drs. Mathew Augustine and Joshua Mendell established a CRISPR-Cas9 library containing 12,474 guide RNAs targeting 660 FDA proved ‘druggable’ putative proteins (18 guides/gene). Using these, the Minna lab performed CRISPR-cas9 library based LOF screening in combination with sub-lethal doses of taxane in in vitro as well as in vivo non small cell lung cancer (NSCLC) models to search for taxane sensitizers. Again we wanted to identify targets that would work in tumor cells growing in vivo. We investigated their potential to chemosensitize (“drop out” in the presence of taxane) the NSCLC lung adenocarcinoma (LUAD) line NCI-H2009 (TP53 and KRAS mutant) in parallel studies in vitro (tissue culture) & in vivo (xenografts). Key elements of our experiments were: the use of low doses of paclitaxel (IC10 values for in vitro and in vivo doses that barely affected tumor growth) compared to control treatment; treatment schedules in vitro and in vivo that mirrored those used in patients; multiple biologic replicates for each transfection and drug selection; and large representation (2,000 cells/sgRNA) for each guide. At the end time point (1 month) we harvested multiple replicates of the taxane and control treatments and sequenced each to identify each guide that “remained” or “dropped out.” We were looking for guides that selectively dropped out in the setting of low dose taxane exposure comparing taxane to control treatment and in vitro to in vivo results. Of importance, we found little overlap between guides that dropped out during taxane exposure in tissue culture vs. those that dropped out in xenografts. From the top 10 gene “sensitizers” (which included FASN, SOAT1, and IMPDH1) that selectively dropped out only with taxane treatment and selectively dropped out in xenografts compared to 2D mass tissue culture, we focused on SOAT1 (Sterol O-Acyltransferase 1). SOAT1 is a key enzyme for lipid metabolism which mediates conversion of intracellular free cholesterol to cholesteryl esters which are then stored as lipid droplets. SOAT1 is a potential cancer therapeutic target with a clinically available inhibitor, avasimibe. H2009 cells with SOAT1 hemizygously removed (CRISPR) grew well in vitro and in vivo in the absence of chemotherapy treatment, but were dramatically sensitized to taxanes compared to parental H2009 cells. While avasimibe treatment, at concentrations achievable in patients, sensitized NSCLC to taxanes, this sensitization was not as dramatic as hemizygous removal by CRISPR indicating the need for additional SOAT1 inhibitors. Of equal importance, SOAT1 H2009 hemizygous knockout (KO) cells were also dramatically sensitized to etoposide, pemetrexed, and gemcitabine, other chemotherapy agents used in NSCLC treatment. Mechanistically, RNAseq analysis identified G2/M cell cycle related gene sets as enriched in the H2009 SOAT1 KO xenografts compared to SOAT1 wildtype xenografts with paclitaxel treatment. Our results using CRISPR screening in vivo, identified SOAT1 as a critical therapeutic chemosensitizing target requiring only 50% inhibition of activity for sensitizing NSCLC to several chemotherapy agents routinely used in the clinic.
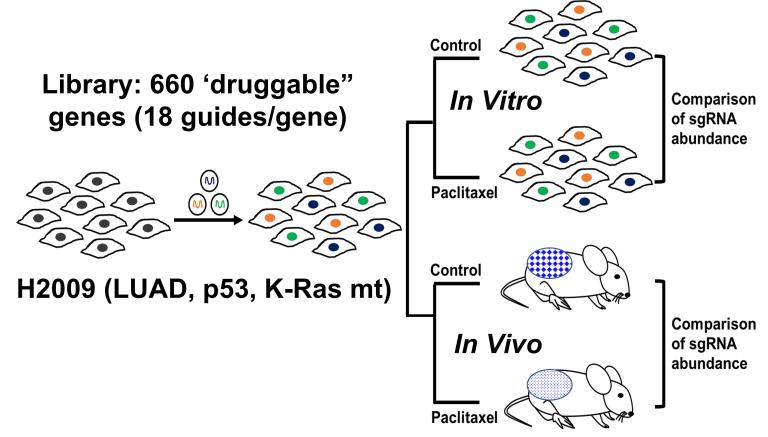
Functional genomics CRISPR screen to identify taxane sensitizers in the “druggable” genome.
Tissue culture and tumor cells are sequenced to determine which guide RNAs have “dropped out” and comparison is made between control and very low dose taxane treatment, and between in vitro and in vivo results. This approach identified a series of druggable proteins (such as SOAT1) that when depleted dramatically sensitize LUAD cells to taxanes but only in the in vivo setting.