Overview
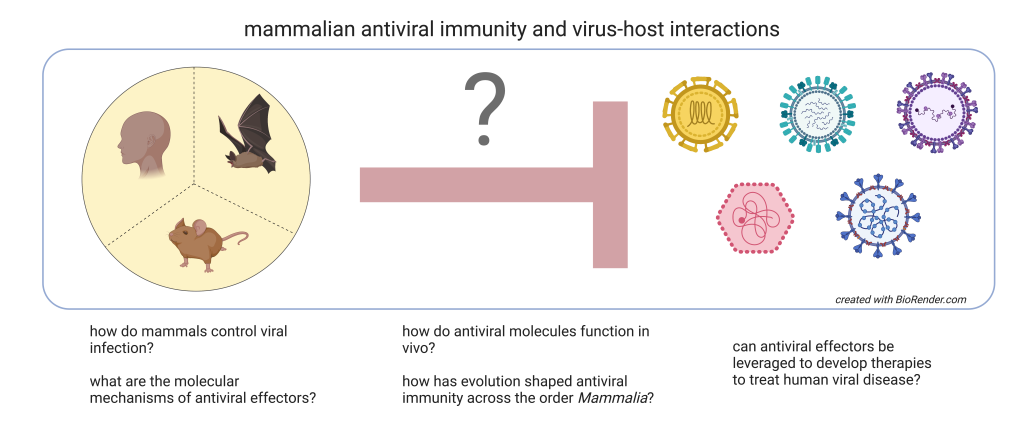
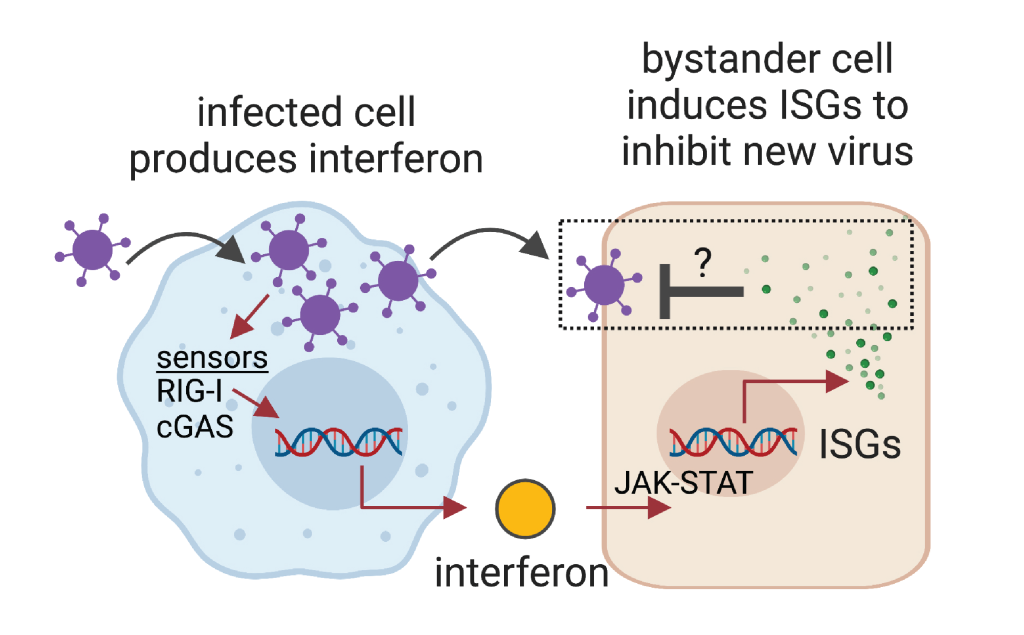
Mammals, humans included, are quite good at fighting off most viral infections by simply relying on their own immune systems. There are exceptions to this, but in general, mammals have evolved a robust antiviral response that ensures their survival when faced with a viral threat. A key component of this response is the interferon pathway. When a cell is infected by a virus, it will produce interferon in response to sensing the viral components. Interferon is secreted from the infected cell and acts to warn neighboring cells to mount their antiviral defenses in the event the virus comes their way. The neighboring cells do this by responding to the interferon stimulus, specifically by initiating de novo gene expression of hundreds of so-called "interferon-stimulated genes". Many of the protein products encoded by these inducible genes have direct antiviral effector properties that can inhibit the invading viruses. Our lab is broadly interested in identifying antiviral effectors, characterizing their specific mechanisms of action, determining their roles in vivo, and studying the evolution of these effectors across diverse mammals (primates, rodents, bats, carnivores, ungulates, etc.)
Our Approaches
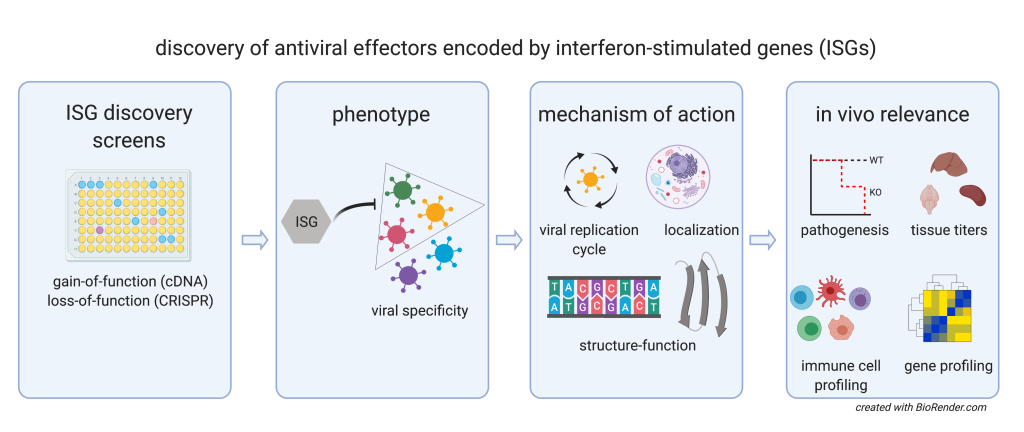
We use two types of genetic screens to uncover antiviral ISGs: 1) cDNA-based ectopic expression to identify genes that, when overexpressed, are sufficient to inhibit viral infection, and 2) CRISPR-Cas9 based gene silencing to identify genes that make significant contributions to the interferon-induced antiviral state. The screens are performed in cell culture lines from diverse mammals (e.g. human, mouse, bat).
Once we identify an antiviral ISG using these screening methods, we then characterize the phenotype of the gene of interest. We determine whether it narrowly suppresses one class of virus, or whether it inhibits broader, more diverse types of viruses. The lab has a large collection of viruses to examine ISG specificity, including flaviviruses, alphaviruses, coronaviruses, picornaviruses, paramyxoviruses, influenza viruses, and herpesviruses.
We then determine the ISG effector mechanism of action. This entails diverse approaches spanning molecular virology, biochemistry, and cell biology. The goals are to assess where the effector is localizes in the cell, which step in the viral replication cycle the effector targets, and how it engages either the virus or other host cell machinery to suppress the virus.
For the most compelling ISGs, we then ask whether the effector contributes to immune protection from viral infection in vivo. If the mouse ortholog is also antiviral, we generate knockout mice that lack the ISG interest. These mice are challenged with a virus that we know should be suppressed. We then monitor the mice for signs of disease and viral replication.
Parallel to this pipeline, we are interested in examining the evolutionary forces that underlie ISG antiviral mechanisms. As a field, we are learning that even though ISG effectors are frequently conserved across Mammalia, they may not retain their antiviral specificity from one species of animal to the next. As a generic example, "ISG A" from bats may inhibit "flavivirus X" but not "flavivirus Y", whereas the human ortholog "ISG A" inhibits "flavivirus Y" but not "flavivirus X". These phenotypes point to the possibility that at the level of antiviral effectors, mammals and viruses are in a constant "arms race" – in order to survive, each side is forced to adapt to the evolutionary pressure imposed by the other side. We seek to understand these virus-host interactions over the 100 million years of mammalian evolution.