Computational approaches to discover mechanisms of somatic and germline mutagenesis
Germline mutations fuel evolution and cause inherited diseases, while somatic mutations underlie clonal expansions, cancer, and likely contribute to aging.
In the Seplyarskiy lab, we use large-scale sequencing data and population-genetic modeling to discover and characterize the mutational mechanisms shaping both germline and somatic genomes. Our work focuses on extracting mechanistic insight directly from patterns of genomic variation, using computational and data-driven approaches.
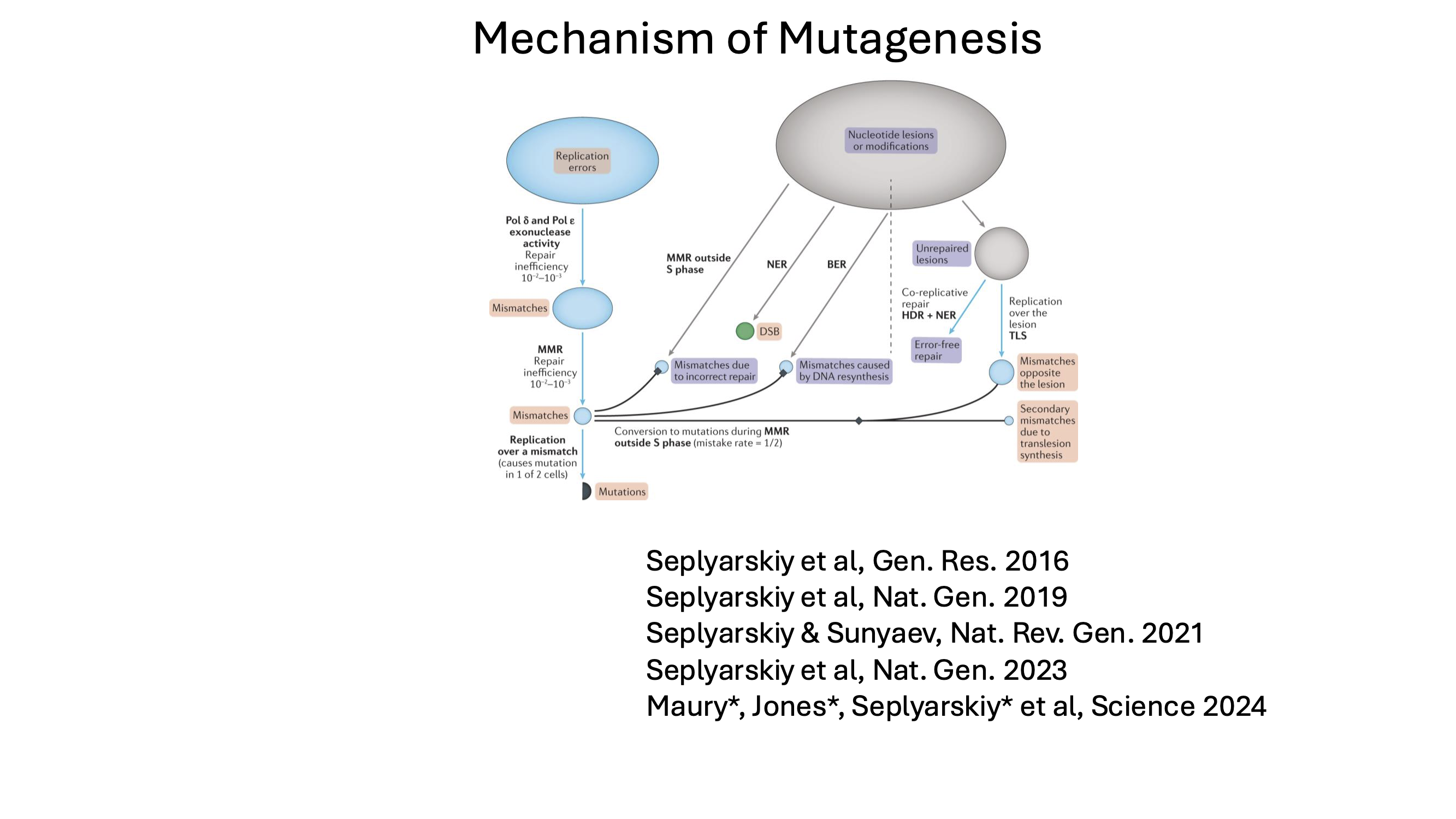
This research direction builds on discoveries that revealed how major mutagenic processes operate in vivo, including how endogenous mutagens gain access to nuclear DNA and how bulky DNA lesions are resolved during DNA replication. These studies showed that distinct mutational mechanisms leave interpretable, quantitative signatures in genomic data.
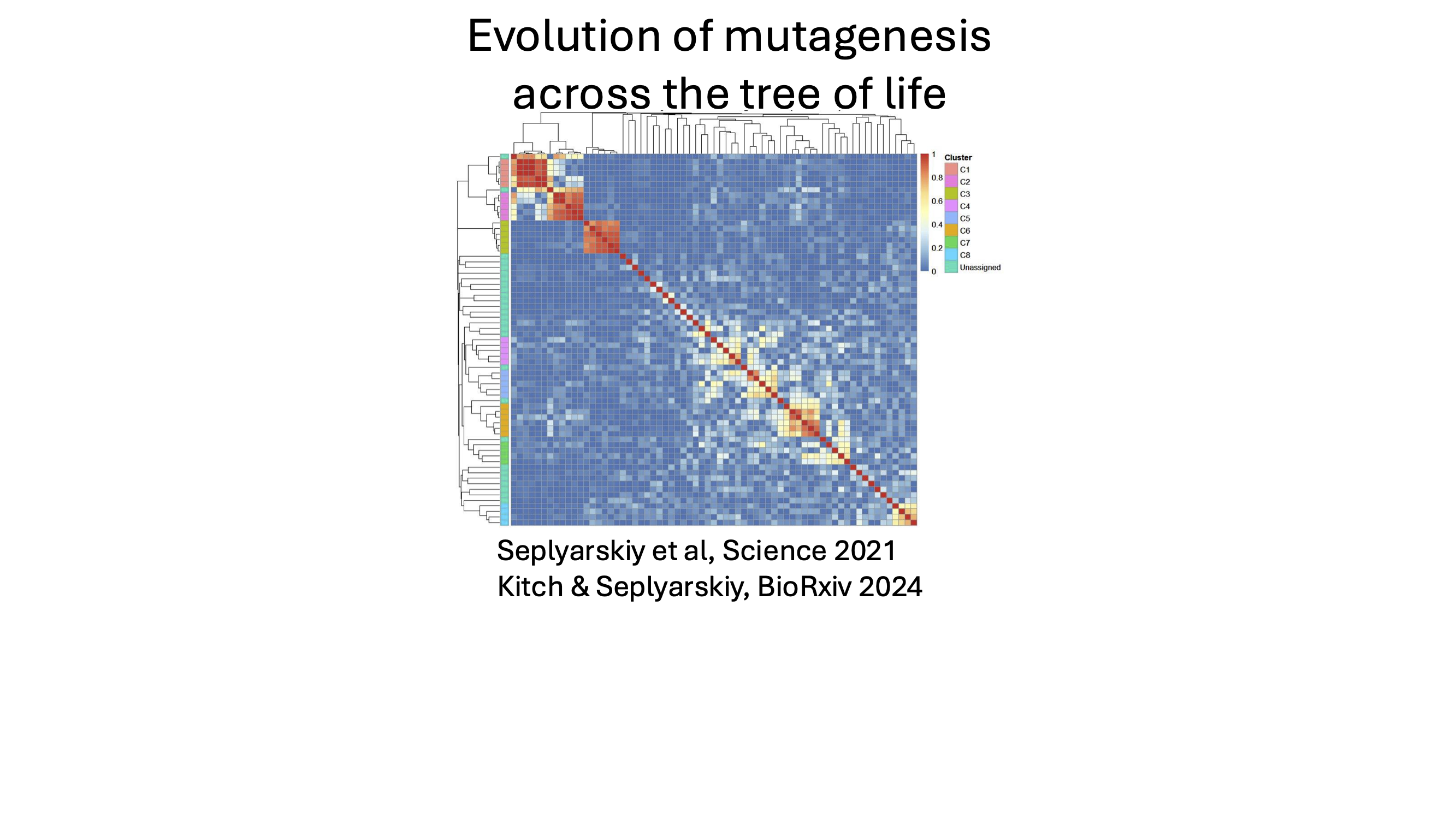
My lab aims to identify previously unrecognized mutational processes, quantify their timing and strength, and characterize their evolution across the tree of life. This knowledge will help elucidate how these processes interact with selection during clonal evolution, cancer development, and aging. A central goal is to develop general computational frameworks that connect observed mutation patterns to underlying molecular mechanisms across tissues and evolutionary timescales.
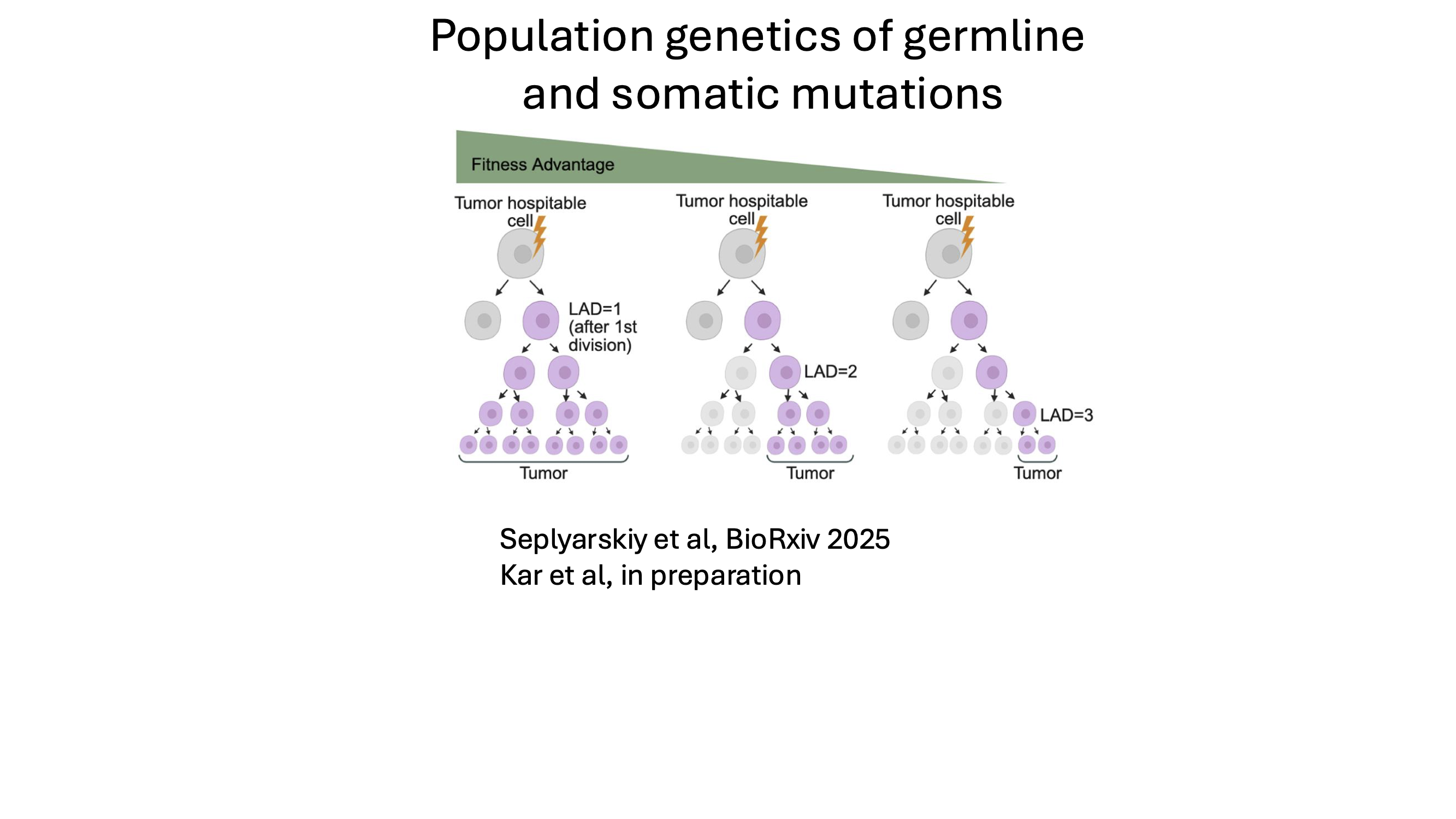
Effect of germline and somatic mutations through population-genetic inference
Functionally important germline mutations affect organismal fitness, while functional somatic mutations can drive clonal expansions within tissues. Despite their different biological contexts, both processes are governed by the same population-genetic principles of mutation, selection, and drift.
In this research direction, we use allele frequency distributions in large genomic datasets to infer the strength and timing of selection acting on germline and somatic mutations. By modeling how mutations propagate in populations and within tissues, we aim to distinguish neutral variation from mutations under positive or negative selection and to connect these signatures to disease risk and functional impact.
Role of de novo mutations in rare disease
A substantial fraction of cases of rare, severe diseases, such as neurodevelopmental disorders, can be attributed to de novo mutations, DNA changes that are absent in healthy parents but present in affected offspring.
Our lab studies the distribution of de novo mutations in large family-based cohorts to estimate the effects of these mutations on phenotype. We also aim to identify hypermutable genes that disproportionately contribute to disease risk. This work directly connects disease genetics to underlying mutational mechanisms and variation in mutation rates across the genome.
Collaborators

We collaborate with Donate Weghorn on mutagenesis.

We collaborate with Evan Koch on population genetics and germline mutagenesis.

We collaborate with Diane Shao on somatic evolution.

We collaborate with Joshua Schraiber and Matt Pennell on evolution of mutational processes on the tree of life.