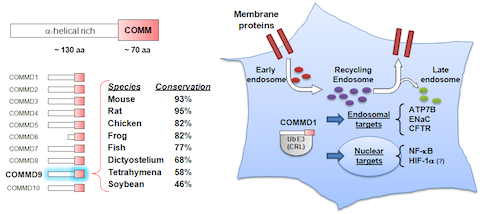
Our initial work led us to the discovery of the COMMD protein family, which is defined by a unique domain (JBC, 2005). Nearly at the same time as our work implicated COMMD1 in immune regulation, genetic studies identified this factor as playing a central role in copper metabolism, through unclear mechanisms. We discovered that COMMD1 regulates cellular copper through a previously unknown protein complex that controls recycling of proteins from the endosomal compartment – we identified and reported the existence of this complex known as the CCC protein assembly (MBoC, 2015). Thereafter, we identified that CCC regulates a novel cargo recognition system that we named Retriever (Nat Cell Bio, 2017) and most recently we described that CCC works through the regulation of endosomal levels of phosphoinositide-3-phoshate (Nat Comm, 2019). Building on these discoveries, we were able to produce a high-resolution structure of Retriever, and an AlphaFold Multimer molecular model for its interaction with CCC (Nat Struct Mol Bio, 2024). This work is supported by NIH R01DK107733, and pending additional support from NIH R01HL179813.