Organelles are subdivided into functionally distinct sub-domains
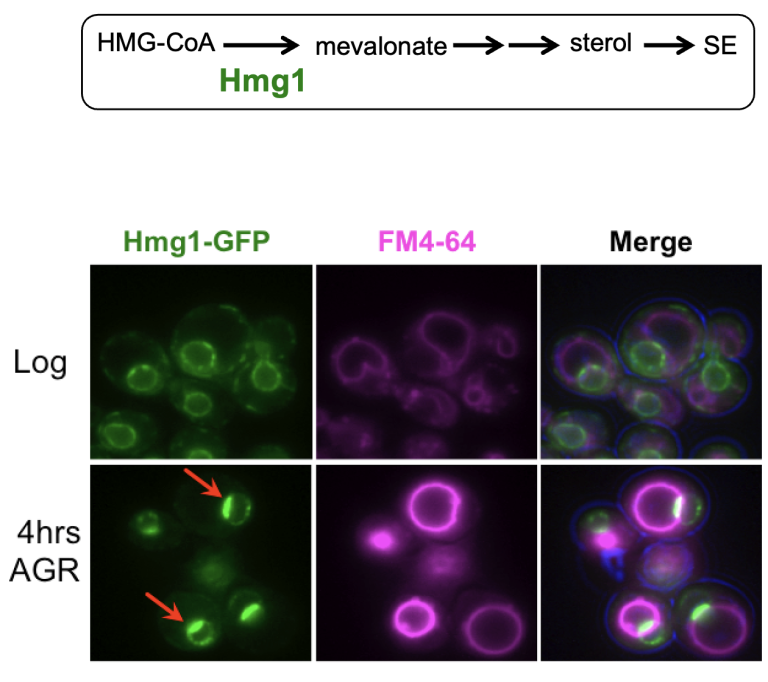
We are interested in how organelles can be functionally subdivided into distinct functional zones. In the past we used budding yeast to study this, and found that the endoplasmic reticulum (ER) can compartmentalize the rate-limiting enzyme of mevalonate biosynthesis, HMG-CoA Reductase (Hmg1), in an ER sub-domain called the nucleus-vacuole junction (NVJ). Student Sean Rogers found Hmg1 partitioning promoted its enzymatic activity, driving mevalonate production so cells could adapt to glucose starvation (Rogers, ELife, 2021). See highlight in ELife here.
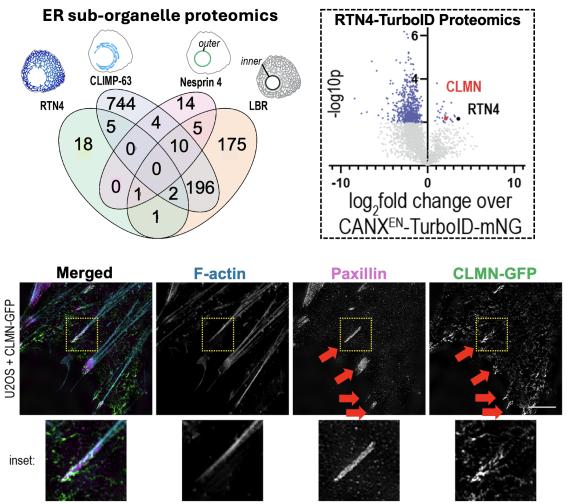
Sub-organelle proteomics mapping reveals ER-actin tether Calmin
Recently, we developed TurboID-based proximity proteomics methods to map the sub-organelle proteome of the ER network. Using CRISPR-Cas9 to fuse TurboID to several ER proteins partitioned into ER sub-domains, we identified dozens of proteins compartmentalized into different ER zones (Merta, Cell Reports, 2025). Among these was a poorly understood protein called Calmin (CLMN), which was highly enriched in ER tubules. We find CLMN is an ER-actin tether that influences focal adhesion disassembly and cell motility.