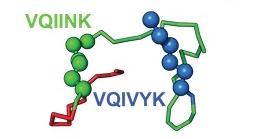
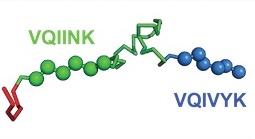
Models of inert tau monomer (Mi, top) and seed-competent monomer (Ms, bottom) suggest that differences in the R1R2 and R2R3 repeat domains underlie the functional difference between the two forms. The VQIINK and VQIVYK sequences drive the formation of amyloid fibrils and are flanked by peptide motifs that form hairpin turns.
We use a variety of analytical approaches to uncover the complex structure-function relationship among intrinsically disordered proteins:
- Cross-linking mass spectroscopy (XL-MS)
(defines contact points within a protein and provides vital clues about its architecture) - Structural analysis
- NMR
- X-ray crystallography
- Cryo-electron microscopy
- Computational methods of protein-structure prediction (e.g., Rosetta method)
- Biochemical methods (e.g., binding assays).
This hybrid structural analysis led to a recent major finding, that structural changes in tau underlie whether or not it aggregates into pathogenic amyloid fibrils. Our initial XL-MS results provided structural information that constrained feasible structures using the Rosetta method of structure prediction. Kinetic analysis further refined the models.
References
Chen, D, et al. (2019) Tau local structure shields an amyloid-forming motif and controls aggregation propensity. Nat Comm 10, 2493.
Mirbaha H, et al (2018). Inert and seed-competent tau monomers suggest structural origins of aggregation. eLife:e36584.