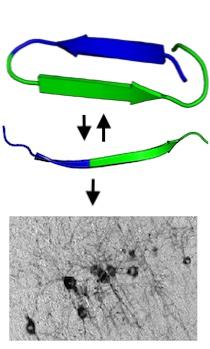
Top: Tau protein can reversibly change conformation to hide and expose the 6-peptide amyloid motif (blue) that plays a critical role in pathological aggregation. Once exposed, the amyloid motif drives the irreversible formation of tangles in brain tissue (bottom).
Traditionally, proteins have been regarded as rigid structures, but recent research has shown that some proteins are highly flexible in both shape and function.
As these intrinsically disordered proteins (IDPs) morph into various shapes, they hide or reveal local regions that are biologically active. Several disease-related proteins, including tau, are IDPs.
We have recently demonstrated that disease-causing mutations in tau protein localize near its "amyloid motif" where the mutations perturb local secondary structure and drive aggregation.
We have also found that the six-amino-acid motif becomes physically inaccessible and biologically inactive in some conformations of tau.
We hypothesize that sequences near the amyloid motifs inhibit protein aggregation, and that these elements play essential roles in controlling aggregation properties of IDPs.
Our lab uses a combination of bioinformatics, structural dynamics, and biophysical techniques to study which shapes of tau prevent or allow pathological aggregation, and to identify the key local regions within tau.
Our ongoing work is to identify which elements within tau prevent amyloid formation, and determine what features are necessary to stabilize these non-aggregating forms.
References
Chen, D, et al. (2019). Tau local structure shields an amyloid-forming motif and controls aggregation propensity. Nat Comm 10, 2493.