Guanosine tetra- and penta-phosphate–collectively known as (p)ppGpp–was discovered in the 1960s as a secondary messenger that signals starvation in bacteria. Increased (p)ppGpp levels slow down bacterial growth, while massively enhancing a bacterium’s tolerance to antibiotics. The ability to produce (p)ppGpp when needed is required for persistent infections of many pathogens.
But even after 50+ years, many puzzles remain about (p)ppGpp signaling. The Wang Lab focuses on identifying targets and mechanisms of (p)ppGpp signaling.
Target Identification
First, (p)ppGpp’s targets vary across bacterial lineages, even though (p)ppGpp itself is universally conserved in the bacteria kingdom. Unbiased target identification has not been performed in most bacterial phyla, so we have no clue how the bacterial starvation response works in some species that substantially affect human health, such as Mycobacterium tuberculosis and Bacteroides thetaiotaomicron.
In the Wang Lab, we use photo-crosslinking probes to capture protein targets of (p)ppGpp for identification using mass spectrometry.
Metabolite-Protein Interactions
Second, growth inhibition by (p)ppGpp requires coordinated down-regulation of essentially all major cellular processes (see figure). Surprisingly, no (p)ppGpp-binding proteins have been identified in several key processes, including DNA replication, fatty-acid synthesis, and cell-wall synthesis.
We hypothesize that processes directly inhibited by (p)ppGpp can crosstalk to other processes via hidden metabolite-protein interactions, and we seek to identify such mechanisms using biochemical and metabolomic approaches.
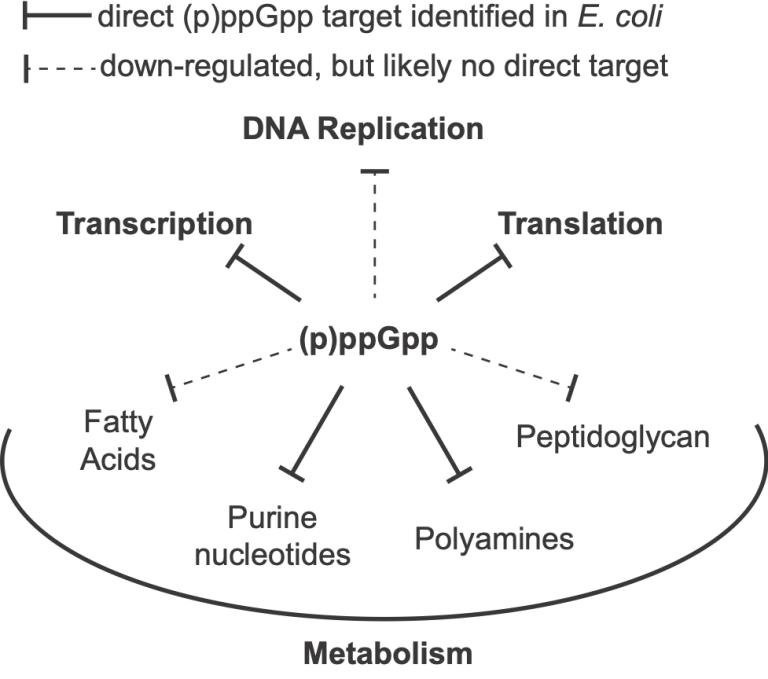 (p)ppGpp affects many cellular processes but has few identified targets. We hypothesize that metabolites sensitive to (p)ppGpp induction are responsible for those elusive inhibition mechanisms.
(p)ppGpp affects many cellular processes but has few identified targets. We hypothesize that metabolites sensitive to (p)ppGpp induction are responsible for those elusive inhibition mechanisms.