Riboswitches – short segments of mRNA that bind to small molecules and cause a change in expression of proteins coded by the RNA – allow protein expression to self-regulate. Few riboswitches have been identified: E. coli, for instance, contains thousands of genes and hundreds of metabolites, but only nine riboswitches have been identified.
The binding sites, or aptamers, are thought to be located in 5’-untranslated regions of mRNA. As a result, computational pipelines for riboswitch discovery have focused on these intergenic regions, overlooking the vast majority of sequences in a transcriptome.
Recent high-resolution footprinting approaches (e.g., DMS-Seq, SHAPE) have shown that the bacterial transcriptome, including coding regions, is highly structured. We, therefore, plan to explore whether this rich structural landscape is recognized by small molecule metabolites for regulatory purposes.
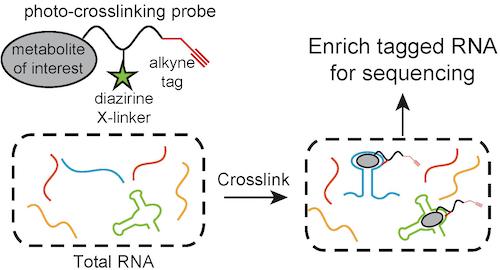
We will use photo-crosslinking probes to capture metabolite-binding RNA for analysis by Illumina sequencing. If successful, this would become the first unbiased approach to identifying metabolite-RNA interactions, and we might discover aptamers outside the current paradigm of riboswitches.