Site-specific functionalization of proteins in vivo lays the fundamentals for numerous powerful chemical biology tools to elucidate protein functions. For example, an enzyme derivatized with a photo-crosslinking group allows capture and identification of its interaction partners, while a protein bearing post-translational modifications enables direct interrogation of its function under this modified state.
Unfortunately, generating these functionalized proteins in vivo has not been possible in most bacterial lineages. There are two popular approaches to incorporating non-canonical functionalities into a protein of interest: genetic codon expansion and protein semi-synthesis.
Genetic codon expansion has flourished in E. coli, but attempts to extend its application beyond the Enterobacteriaceae family have mostly seen unsatisfactory efficiency or specificity.
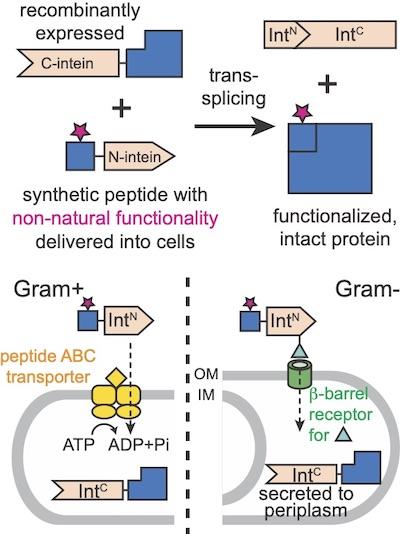
We are therefore interested in implementing the protein semi-synthesis approach (top panel) in major model bacterial organisms other than E. coli. The key to the success of this approach is the efficient delivery of a synthetic peptide, 20-50 residues in length, across the bacterial cell membrane or envelope (bottom panel). Thus, we will start this project by developing peptide delivery machinery through directed evolution.