Ribosome biogenesis
All protein synthesis depends on ribosomes. Disruption of ribosome production or turnover has important functional consequences: enhanced ribosome biogenesis drives tumor growth, inflammation, and hypertrophy, while defects in ribosome assembly and function cause a variety of developmental disorders, age-related phenotypes, and neurodegeneration.
Over 200 proteins have dedicated roles in ribosome biogenesis, providing many potential druggable targets. Despite the therapeutic possibilities, few small molecules that promote or inhibit specific steps in ribosome assembly have been identified. The major obstacle to identifying new modulators of ribosome production has been the absence of a platform amenable to large-scale screening.
To overcome this significant barrier, we have generated several powerful pulse-labeling reporters (ribo-SNAP reporters) that allow us to track heterochronic ribosomes within human cells at single cell resolution in real time. Using these tools, we have gained new insights into the dynamics of ribosome assembly in different cell types under a variety of experimental conditions. These include cancer cell lines, human induced pluripotent stem (iPS) cell lines, and neurons and cardiomyocytes derived from iPS cells. We are now in a unique position to screen for small molecules and mutations that enhance or inhibit ribosome production in a variety of contexts.
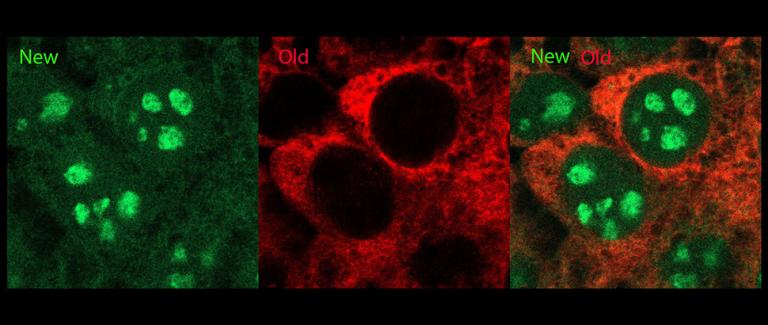 Chronologically distinct ribosome populations labeled in the same living cell. Red indicates old ribosomes; green indicates new ribosomes.
Chronologically distinct ribosome populations labeled in the same living cell. Red indicates old ribosomes; green indicates new ribosomes. rRNA transcription
Most research on ribosome biogenesis and protein synthesis uses yeast or mammalian cell lines, but this work provides only an average picture of these fundamental processes; it can't reveal dynamic differences among individual cells in vivo.
Strikingly, emerging data show that the dynamic regulation of rRNA transcription and ribosome biogenesis directly impacts the fate and function of different cell types across species.
Unfortunately, robust and genetically tractable models for studying the differential regulation of ribosome formation have not been available until recently. We overcame this obstacle by identifying a Drosophila RNA Polymerase I (Pol I) regulatory complex that resembles the human SL1 complex. This complex regulates the proliferation and growth of germline stem cells (GSCs). Our published work and new preliminary data indicate that modulation of Pol I activity influences cell fate and function in several different tissues.
A critical question now becomes how different types of cells establish different rates of rRNA transcription and ribosome assembly. Our discovery of the Drosophila SL1 complex provides a unique opportunity to identify and characterize mechanisms that modulate rRNA transcription and ribosome biogenesis in different cell types in vivo. We have also begun to characterize several enzymes, including NO66 and MINA, which may regulate ribosome biogenesis and function.
To explore how and what regulates rRNA transcription, we are seeking to:
- Reconstitute rRNA transcription reporter in mammalian cells
- Genome-wide screening to find positive and negative regulators of rRNA transcription
- rRNA transcription changes in various stress conditions
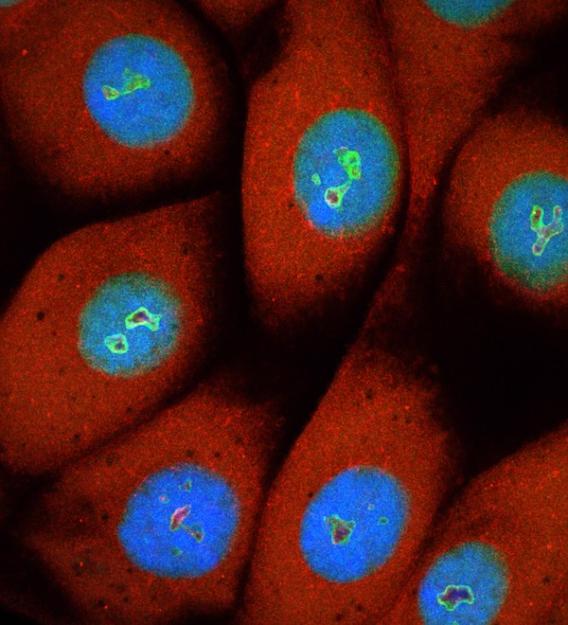 Stained salivary-gland cells. Nuclei are stained in blue; green indicates nucleoli, the site of ribosome biogenesis.
Stained salivary-gland cells. Nuclei are stained in blue; green indicates nucleoli, the site of ribosome biogenesis.