My lab has a long-time interest in understanding the mechanisms of transcription and gene regulation in mammalian cells using initially cell-free systems reconstituted with purified gene-specific transcription factors, general cofactors, and components of the general transcription machinery to recapitulate transcriptional events in vitro.
Mechanistic studies based on cell-free transcription systems with DNA and chromatin templates are further evaluated in vivo by chromatin immunoprecipitation (ChIP), RT-PCR, reporter gene assay, co-immunoprecipitation (co-IP), nucleosome mapping, siRNA/shRNA knockdown, and CRISPR-Cas9 knockout using different types of cultured cells.
Recently, we have also adapted conditional knockout and transgenic knock-in mice to investigate the functional role of bromodomain-containing protein 4 (BRD4) using both xenograft and syngenic cancer models. In addition, patient-derived xenografts (PDXs) and organoids are used to evaluate BRD4 protein isoforms in cancer initiation and progression.
Our goals are to elucidate the general principles underlying gene activation and repression in mammalian cells and their associated viruses, in particular, DNA tumor viruses such as human papillomaviruses (HPVs).
The application of reconstituted chromatin transcription systems and HPV and cancer models has significantly advanced our mechanistic understanding of eukaryotic gene transcription and chromatin dynamics.
Broadly, our research is categorized in these six areas:
Role of Human General Transcription Factors and Cofactors in Eukaryotic Transcription
Several general transcription factors, including TFIIB, TFIID, TFIIE, TFIIF, and TFIIH, are essential for transcription on most eukaryotic promoters by RNA polymerase II (Pol II).
The core promoter-binding factor TFIID, which is comprised of the TATA-binding protein (TBP) and a dozen or so evolutionarily conserved Pol II-specific TBP-associated factors (TAFs), has an intrinsic activity to recognize the TATA box, initiator, and downstream promoter elements and initiates preinitiation complex assembly on both TATA-containing and TATA-less promoters. TFIID also functions as a general cofactor in transducing regulatory signals to the general transcription machinery and plays a crucial role in transcription from chromatin templates due to multiple enzymatic activities inherent to its TAF components.
These diverse features have implicated TFIID as a central player in eukaryotic transcription and as a core promoter-defining factor.
To dissect the mechanisms of gene activation and repression in mammalian cells, we have employed human cell-free transcription systems reconstituted either with:
a) individually purified general transcription components (TFIIA, TFIIB, TBP, TFIIE, TFIIF, and PC4 coactivator) and highly purified epitope-tagged protein complexes (TFIID, TFIIH, and pol II)
or
b) with TFIID and a preassembled Pol II holoenzyme complex that contains components of the general transcription machinery as well as SWI/SNF chromatin remodeling factor, GCN5 histone acetyltransferase, and Mediator complex.
Our objectives are to define the biochemical activities of general transcription components in modulating gene activity and to uncover the combinatorial nature of eukaryotic gene regulation using well-defined cell-free transcription systems reconstituted with purified human proteins.
Transcriptional Regulation and DNA Replication in Human Papillomaviruses (HPVs)
HPVs induce many human diseases, including skin warts, genital warts, cervical cancer, anal cancer, and head-and-neck cancer.
We are interested in understanding the mechanisms by which virus-encoded E2 and E6 proteins activate or repress HPV transcription in the context of both DNA and chromatin templates.
Using in vitro-reconstituted HPV mini-chromosome that faithfully recapitulates in vivo phasing of HPV chromatin, we first identified bromodomain-containing protein 4 (BRD4) as the cellular adaptor mediating the repressing activity of HPV E2 that controls E6 and E7 gene transcription.
We have elucidated the repression mechanisms employed by the E2-BRD4 silencing complex and defined the role of SMC5 and SMC6 proteins in regulating E2 function in transcription, cell cycle checkpoint control, viral DNA replication, and HPV genome maintenance and segregation.
HPV-encoded E6 inhibits p53 target gene transcription by inducing p53 degradation through ubiquitin-dependent proteosome pathway or by epigenetic mechanisms through suppressing of p300/CBP histone acetyltransferase (HAT) activity independent of p53 degradation pathway.
Bromodomain-Containing Protein 4 (BRD4)
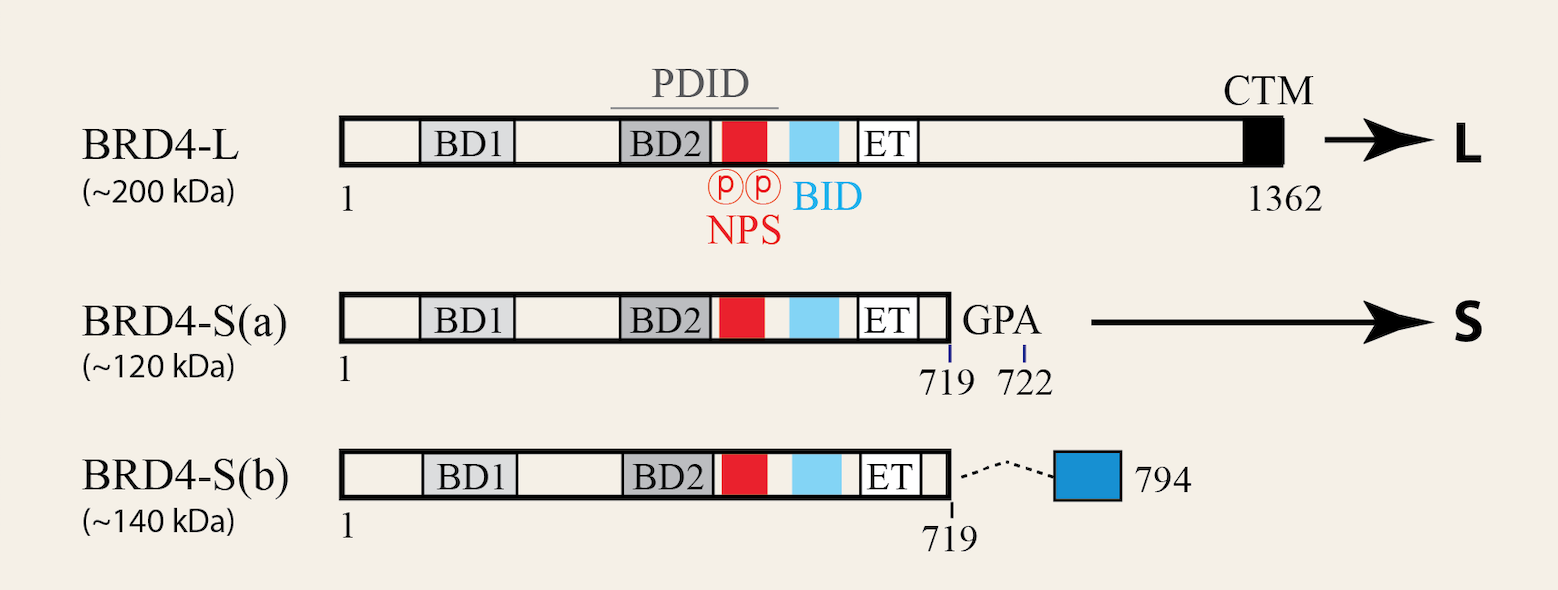
Our lab has identified human bromodomain-containing protein 4 (BRD4) as a crucial epigenetic regulator in viral and cellular transcription programming and chromatin dynamics, including DNA replication and genomic maintenance. Since our first description of BRD4 as an HPV E2 corepressor in suppressing HPV transcription (2006 G&D paper), we have continued to elucidate the functional and biological roles of BRD4 in various biological systems through our lab research and also collaborative work with scientists and laboratories around the world.
See our research on BRD4-Regulated Transcription Programs and Cancer Pathways and Therapeutics Targeting Phospho-BRD4
Post-translational Modification
Our lab has been working on the activation and repression mechanisms by which p53 and AP-1 modulate cellular and viral gene expression via:
-
functional association with BRD4 and general transcription components.
-
how posttranslational modifications (methylation, acetylation, sumoylation, neddylation, and ubiquitination) fine-tune the transcriptional activity of these transcription factors via covalent linkage of critical lysine residues.

p53 Post-translational Modifications

Crosstalk Between Acetylation and Sumoylation Regulates p53 Binding to DNA
Reconstituted histone acetyltransferase (HAT) assays and in vitro methylation, phosphorylation, sumoylation, neddylation, and ubiquitination assays are routinely used in the lab to define the role of posttranslational modification in modulating transcription factor activity and various signaling pathways.
We are also interested in:
dissecting the molecular mechanism underlying p300/CBP and G9a/GLP involvement in gene activation and repression by switching its role from activation to repression
and
how phosphorylation regulates BRD4 chromatin targeting and recruitment of various transcription factors and chromatin modifiers, with concurrent development of phospho-BRD4-targeting compounds to modulate these processes.
Transcription Programming and Cancer Pathways
BRD4-Regulated Transcription Programs and Cancer Pathways
The identification of BRD4 as a key transcription regulator in HPV and cellular gene transcription, DNA replication, DNA repair, cell cycle control, and stem cell renewal and differentiation has prompted my lab research in BRD4-centered gene regulation and chromatin dynamics over the past 15 years.
Our interest has progressed from:
- transcription mechanisms
- phosphorylation-driven BRD4 gene control and factor recruitment
- loss and gain of p53 functions
- DNA replication
- mitotic chromosome progression and bookmarking
- DNA repair
to:
- cancer etiology
- cellular systems
- genome editing
- in vivo cancer models
Our initial interest in HPV-driven cervical cancer and head-and-neck cancer has been further expanded to non-HPV-associated breast cancer and other human diseases. Recent studies have focused primarily on how phosphorylation controls BRD4 protein conformation, factor recruitment, and pathway selectivity and also how BRD4 protein isoforms modulate cancer initiation and progression. The discoveries we have made over the years continuously shed new light on the roles of BRD4 in viral and cellular processes and disease implications.
Therapeutics Targeting Phospho-BRD4
The identification of BRD4 as a cancer therapeutic target through pathway discovery and development of small compound inhibitors and PROTAC-based BRD4 degraders has significantly advanced research interest in BRD4.
Our lab has identified dozens of small compound inhibitors targeting phospho-BRD4 through high-throughput compound library screening.
We are currently refining these compound studies and hoping to develop more potent and selective isoform-specific inhibitors and/or degraders to inhibit only oncogenic but not the tumor-suppressive activity of BRD4, allowing more precise targeted therapy for future cancer patient treatment.