In all of our projects, the first step on the way from microscopy images to quantitative understanding of the studied system is to computationally analyze the acquired images. Computational image analysis has the advantages of being efficient and consistent, quantitative and complete, and, when combined with advanced data analysis, may reveal complex, even invisible, relationships within the studied system.
Computer vision challenges that are relevant to our ongoing projects include:
Low-contrast cell segmentation
In many of our studies, we must segment the cell to provide cellular context for the imaged molecules. Yet, some of our cell images look like this:
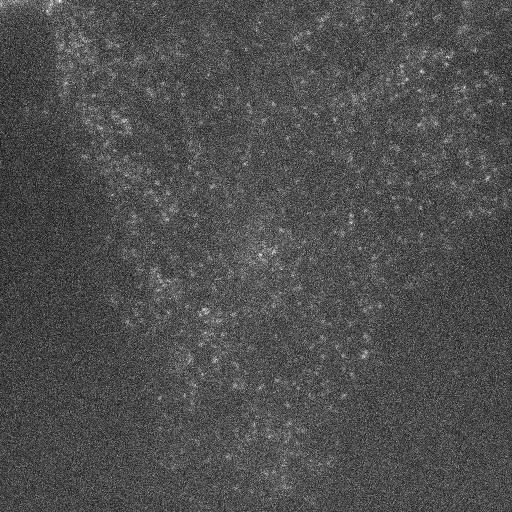
To address this challenge, we are exploring approaches that integrate temporal, spatial, and prior information to identify the cell boundary in such low-contrast images.
Line and junction filtering and detection
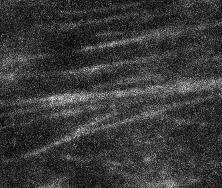
As described on the Biological Questions page, an important subcellular context for membrane proteins is the actin cytoskeleton, which forms overlapping and intersecting linear structures in an image when densely labeled, as shown in the example image to the right.
We are interested in segmenting such actin structures to investigate their link to e.g. receptor or biomolecular condensate behavior. To achieve this, we are developing an algorithm based on high angle-resolution line filtering and multi-angle processing of the filter response to simultaneously detect lines and junctions of arbitrary geometry. (In contrast, existing filters focus on either lines or junctions, but not both together.)
Below is an example application of our algorithm to lamin meshworks at the nuclear envelope, part of a collaboration with the Goldman Lab at Northwestern University Feinberg School of Medicine:
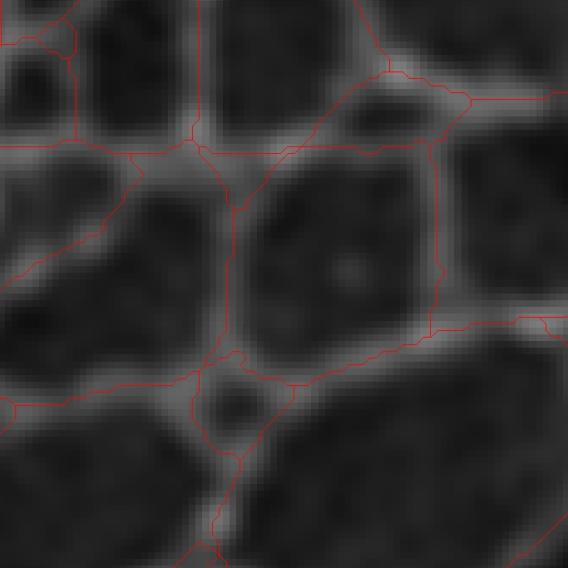
Multiple particle tracking with particle interactions
Almost all of our projects involve particle tracking, for which we use our previously published multiple-particle tracking software (Jaqaman et al. Nature Methods 2008). However, we are always striving to improve our tracking, especially to handle molecular interactions, visualized as merging and splitting events between particles in the image. The challenge in this task is to distinguish between interactions and molecules passing by each other, with the added complexity of fluorophore blinking and imperfect particle detection.