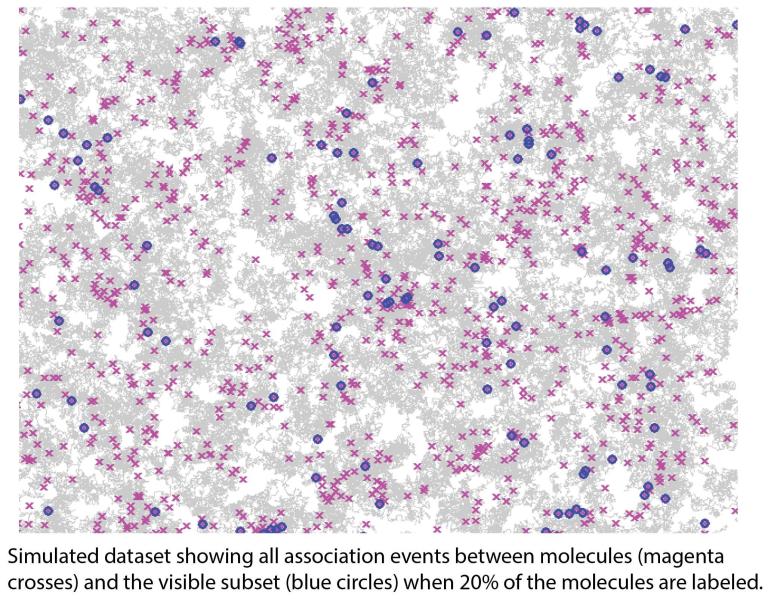
Extracting molecular interaction kinetics from single-molecule data
Live-cell single-molecule imaging has the unique ability to capture discrete molecular interaction events in their native environment. Yet it suffers from the inherent limitation of sparse labeling, i.e. not all molecules of interest are visible at the same time.
The question we are tackling is: Can we extrapolate from the labeled subset to the full population, thus estimating association and dissociation rates between molecules in their native environment? (Can we see the invisible?) In pursuit of this goal, we are taking a mathematical modeling approach. The basis of the approach is to build a stochastic mathematical model that mimics the biological system, e.g. receptor dynamics and interactions, and the data acquisition setup, including sparse labeling, and then calibrate the model with single-molecule data (which are also stochastic).
For this, we are exploring stochastic model calibration strategies, such as Indirect Inference and Approximate Bayesian Computation. We are also looking into GPU programming to speed up our simulations and make the framework practically applicable to experimental data. Our initial tests are very promising and suggest that this framework will eventually have the power to reveal in situ interaction kinetics between molecules. Additionally, we plan to use it to compute multiplex data from different experiments and achieve a bigger-picture understanding of inter-receptor interactions beyond what individual experiments can reveal.