The bacterial flagellum is one of only three known reversible rotary motors produced in nature. Many bacterial species produce a flagellar motor with a commonly conserved core structure that includes the inner membrane-bound secretion system, the flagellar rod and hook, and the extracellular filament. Even though these core flagellar structures are conserved in different bacteria, the functional outputs of these motors can vary across species. For instance, flagellar motors produced by E. coli and Salmonella promote efficient swimming motility in low viscosity environments. However, other bacteria such as Campylobacter jejuni produce flagellar motors that promote swimming motility as efficiently as E. coli in the low-viscosity milieu and also enable augmented motility velocities in higher viscosities that normally paralyze other flagellated bacteria.
C. jejuni swimming motility in low-viscosity media (1 cP).
C. jejuni swimming motility in high-viscous media (40 cP).
Through an exciting collaboration with the laboratory of Dr. Morgan Beeby at Imperial College in London, we are combining traditional approaches such as genetic screens, protein biochemistry, and transmission electron microscopy and innovative technologies including cryo-electron tomography to understand the form and function of flagellar rotary motors across bacterial species, especially flagellar motors that enable high motility velocities through high viscosity environments.
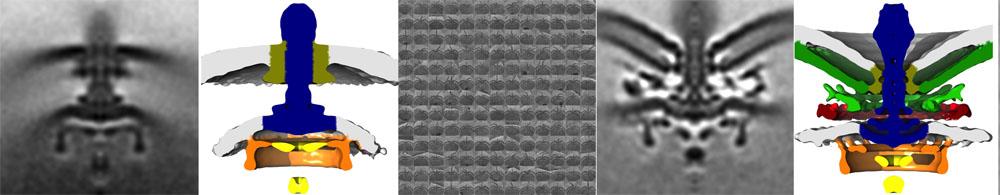 Cryo-electron tomograms of flagellar motors of Salmonella and Campylobacter jejuni
Cryo-electron tomograms of flagellar motors of Salmonella and Campylobacter jejuni Our research indicates that not all flagellar motors are built the same. Indeed, by cryo-electron tomography, we and others have determined average three-dimensional tomograms of intact flagellar motors at low-nanometer resolution and observed variability in flagellar motors structures beyond the core basal body that is common to most flagella.
In C. jejuni, we discovered multiple, large disks composed of different proteins that form on the conserved flagellar core structure, in addition to alterations in the spatial arrangement and dimension of specific flagellar parts. Through transposon mutagenesis, deletion analysis of specific genes, and protein-interactions assays, we are identifying unique proteins required for the structure and function of these complex flagellar motors. Through comparative analysis of tomograms of flagellar motors across species and from isogenic mutants in C. jejuni, we discovered that these additional flagellar structures alter motor function to generate higher torque, which enables high motility velocities in viscous milieus, such as those found in the intestinal mucus layers in hosts. We continue to identify novel flagellar proteins of these high-torque flagellar rotary motors in bacteria and determine how these proteins contribute to the function of these fascinating nanomachines.
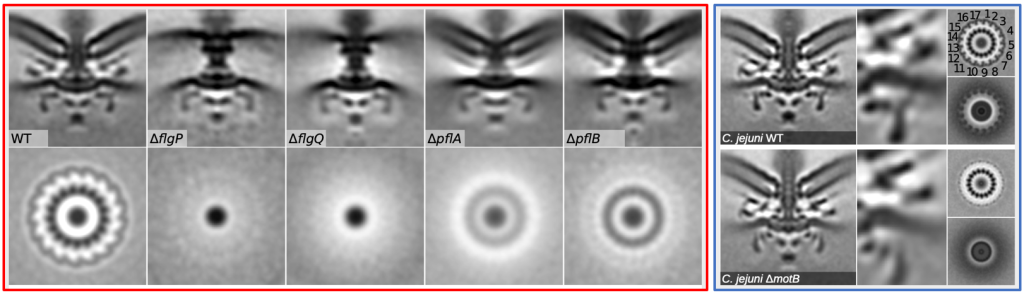 Step-wise generation of flagellar disk structures (left) to incorporate more MotAB stator units (right) into the C. jejuni flagellr motor to generate higher power and torque during flagellar filament rotation.
Step-wise generation of flagellar disk structures (left) to incorporate more MotAB stator units (right) into the C. jejuni flagellr motor to generate higher power and torque during flagellar filament rotation. Relevant Publications
Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility
Hendrixson, D. R., Akerley, B. J., and DiRita, V. J. Mol Microbiol (2001) 40:214-224
Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni
Sommerlad, S. M. and Hendrixson, D. R. J. Bacteriol (2007) 189:179-186
Structural diversity of bacterial flagellar motors
Chen, S., Beeby, M., Murphy, G. E., Leadbetter, J. R., Hendrixson, D. R., Briegel, A., Li Z., Shi, J., Tocheva, E. I., Müller, A., Dobro, M. J., and Jensen, G. J. EMBO J (2011) 30:2972-2981
Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold
Beeby, M., Ribardo, D. A., Brennan, C. A., Ruby, E. G., Jensen, G. J., and Hendrixson, D. R. Proc Natl Acad Sci (2016) 113:E1917-26
A Chaperone for the stator units of a bacterial flagellum
Ribardo, D. A., Kelley, B. R., Johnson, J. G., and Hendrixson, D. R. mBio (2019) 10:e01732-19
Diversification of Campylobacter jejuni flagellar C-ring composition impacts its structure and function in motility, flagellar assembly, and cellular processes
Henderson, L. D., Matthews-Palmer, T. R. S., Gulbronson, C. J., Ribardo, D. A., Beeby, M., and Hendrixson D. R. mBio (2020)11:e02286-10