Campylobacter jejuni is a leading cause of bacterial diarrheal disease in humans in the United States and other developed countries throughout the world. Upon infection of humans, C. jejuni adheres to and invades colonic epithelial cells resulting in inflammation and diarrhea.
 Outcomes of Interactions of C. jejuni with Human and Avian Hosts
Outcomes of Interactions of C. jejuni with Human and Avian Hosts C. jejuni is also a natural commensal organism of intestinal tracts of many animals in the wild and in agriculture, including bovine, porcine, and avian species. The bacterium is highly prevalent in chickens, with the consumption or handling of contaminated chicken meat representing the leading factor in sporadic cases of C. jejuni diarrheal disease. Outbreak cases of diarrheal disease are often due to the consumption of unpasteurized milk or contaminated water sources. C. jejuni can colonize throughout the avian intestinal tract with the highest levels in the ceca and large intestines. C. jejuni often resides in the mucus layer atop the intestinal epithelial cells with little to no invasion across the epithelial layer apparent. As a result, C. jejuni can promote persistent colonization with chickens and other animals without causing any symptoms of the disease.
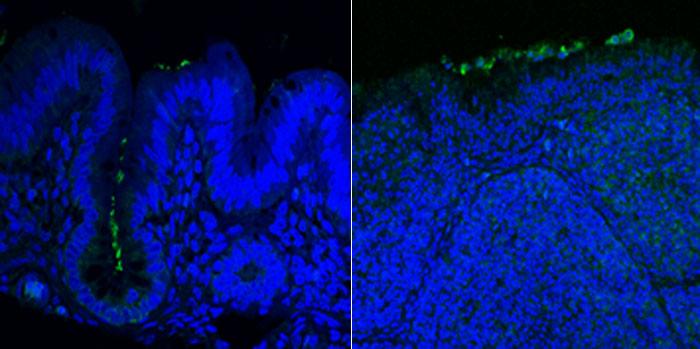 C. jejuni colonization of the avian cecum and bursa
C. jejuni colonization of the avian cecum and bursa One of the goals of our research program is to identify both virulence factors of C. jejuni that allow the bacterium to infect humans to cause diarrheal disease and colonization factors that contribute to the ability of the bacterium to promote harmless, commensal colonization of avian hosts. These types of factors are of great importance as they may represent targets to develop different types of antimicrobials and therapeutics to lessen the ability of C. jejuni to infect humans or reduce the amount of C. jejuni in agriculture and the human food supply.
We have employed different types of strategies to identify factors of C. jejuni that are required for host infection, including negative genetic selections and transcriptome analysis. Through these approaches, we have found roles for specific amino acid transporters, peroxidases, the flagellum, and flagellar motility.
One class of factors we identified include proteins we have annotated as Feds, for flagellar co-expressed determinants. These proteins are transcribed simultaneously with flagellar proteins but are not required for motility. Instead, most of the Feds are required for optimal commensal colonization of the chick intestinal tract or invasion of human colonic cells. Furthermore, some Fed proteins are secreted by the flagellum and likely perform an activity outside the bacterium that is important for interaction with a host.
We are focused on achieving a thorough understanding of why these factors are required by C. jejuni for interactions with a host. In addition, we are engaged in innovative approaches to better understand how C. jejuni interacts with a host and in vivo activities of the bacterium and the host to result in different outcomes of infection.
Relevant Publications
Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract
Hendrixson, D. R., and DiRita, V. J. Mol Microbiol (2004) 52:471-484
Characterization of two cytochrome c peroxidases of Campylobacter jejuni involved in promoting commensal colonization of poultry
Bingham-Ramos, L. K. and Hendrixson, D. R. Infect Immun (2008) 76:1105-1114
Analysis of the LIV system of Campylobacter jejuni reveals alternative roles for LivJ and LivK in commensalism beyond branched-chain amino acid transport
Ribardo, D. A. and Hendrixson, D. R. J. Bacteriol (2011) 193:6233-6243
Identification and analysis of flagellar co-expressed determinants (Feds) of Campylobacter jejuni involved in colonization
Barrero-Tobon, A. M. and Hendrixson, D. R. Mol Microbiol (2012) 84:352-369
Flagellar biogenesis exerts temporal regulation of secretion of specific Campylobacter jejuni colonization and virulence determinants
Barrero-Tobon, A. M. and Hendrixson D. R. Mol Microbiol (2014) 93:957-974
Campylobacter jejuni BumSR directs response to butyrate via sensor phosphatase activity to impact transcription and colonization
Goodman, K. N., Powers, M. J., Crofts, A. A., Trent, M. S., and Hendrixson, D. R. Proc Natl Acad Sci (2020) 117:11715-11726