Functional annotation of cancer genomes through transposon mutagenesis
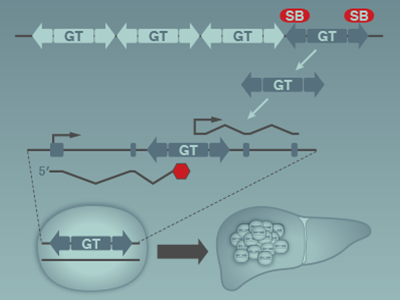
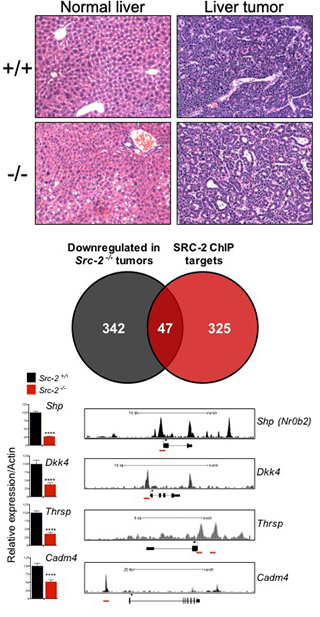
Our group previously utilized the Sleeping Beauty DNA transposon system to identify mutations that cooperate with MYC to accelerate liver tumorigenesis in mice. This led to the identification of Steroid Receptor Coactivator 2 (SRC-2, also known as NCOA2) as a novel gene that functions to restrain MYC-induced liver cancer (O’Donnell et al, PNAS, 2012). SRC-2 encodes a potent transcriptional coactivator that cooperates with nuclear receptors to control multiple physiological processes. Despite evidence from our previous work supporting a tumor suppressor role for SRC-2 in liver cancer, other studies reported an oncogenic role for SRC-2 in other tumor types such as prostate cancer. Thus, additional studies were needed to establish whether SRC-2 is a bona fide cell-autonomous tumor suppressor in liver and to elucidate downstream mechanisms through which it acts as such. In a recent study (Suresh et al., PLoS Genetics, 2017), we provided unequivocal evidence that SRC-2 restrains MYC-mediated liver tumorigenesis in vivo and identified key downstream SRC-2 target genes that mediate this effect. These results provided important new insights into the mechanism of tumor suppression by SRC-2 in MYC-induced liver cancer and have broad implications for our understanding of the functions of nuclear receptor coactivators in cancer.