Many extracellular and intracellular signals culminate at signal-regulated transcription factors in the nuclei of cells. The Kraus Lab is interested in how these signals control transcription factor activity, enhancer formation, and target gene expression in a broad array of biological systems. We are particularly interested in pathways controlled by estrogen receptor alpha (ERα), NF-kβ, and Stat1α. We are also interested in the kinetics of the signal-regulated transcriptional response, and the detailed molecular mechanisms of transcription complex assembly and function. We use a wide range of methodological approaches, from biochemistry to genomics, to study fundamental questions related to normal physiology and disease. The Kraus Lab has leveraged new genomic methodologies, including global run-on sequencing (GRO-seq), to explore signal-regulated transcription in multiple cell types.
Current projects are focused on: (1) enhancer assembly and function, (2) mechanisms of target gene expression in response to cellular signals, and (3) downstream biological outcomes related to cancer, stem cells, inflammation, metabolism, and reproduction.
Our studies are revealing the molecular mechanisms and biological outcomes of various cellular signaling patterns.
Selected Publications
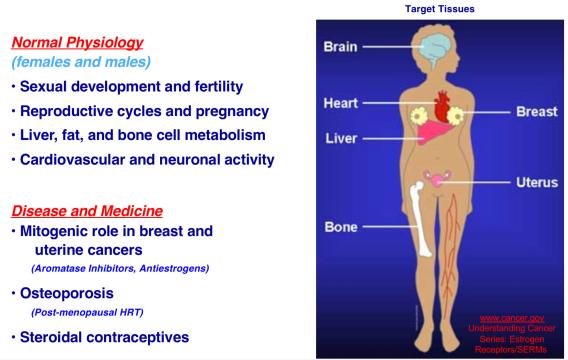
Hah N., Danko C. G., Core L., Waterfall J. J., Siepel A., Lis J. T., Kraus W. L. (2011) A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145:622-634. PMID: 21549415
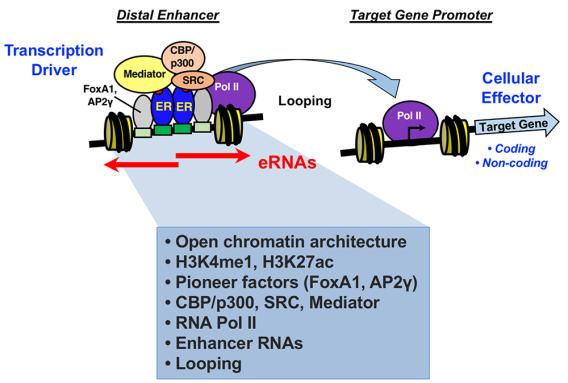
Hah N., Murakami S., Nagari A., Danko C.G., Kraus W. L. (2013) Enhancer transcripts mark active estrogen receptor binding sites. Genome Research 23:1210-1223. PMID: 23636943
Danko C. G., Hah N., Luo X., Martins A., Siepel A., Kraus W. L. (2013) Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Molecular Cell 0:212-222. PMCID: PMC3640649
Luo X., Chae M., Krishnakumar R., Danko C. G., Kraus W.L. (2014) Dynamic reorganization of the AC16 cardiomyocyte transcriptome in response to TNFα signaling revealed by integrated genomic analyses. BMC Genomics 15:155. PMID: 24564208
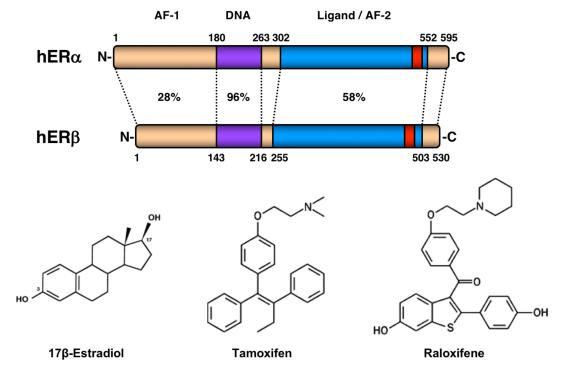
Franco H.L., Nagari A., Kraus W.L. (2015) TNFα signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Molecular Cell 58:21-34. PMCID: PMC25752574
Murakami S., Nagari A., Kraus W.L. (2017) Dynamic assembly and activation of estrogen receptor α enhancers through coregulator switching. Genes & Development 31:1535-1548. PMID: 28887413
Franco H.L., Nagari A., Malladi V.S., Li W., Xi Y., Richardson D., Allton K.L., Tanaka K., Li J., Murakami S., Keyomarsi K., Bedford M.T., Shi X., Li W., Barton M.C., Dent S.Y.R., Kraus W.L. (2018) Enhancer transcription reveals subtype-specific gene expression programs controlling breast cancer pathogenesis. Genome Research 28(2):159-170. PMID: 29273624
Ryu K.W., Nandu T., Kim J., Challa S., DeBerardinis R.J., Kraus W.L. (2018) Metabolic regulation of transcription through compartmentalized NAD+ biosynthesis. Science 360(6389):eaan5780. PMID: 29748257
Hou T.Y., Kraus W.L. (2022) Analysis of estrogen-regulated enhancer RNAs identifies a functional motif required for enhancer assembly and gene expression. Cell Reports 39:110944. PMCID: PMC9246336